Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
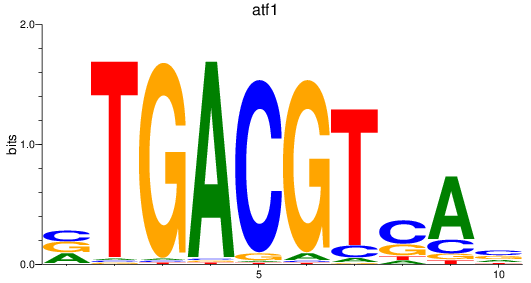
Results for atf1
Z-value: 1.64
Transcription factors associated with atf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf1
|
ENSDARG00000044301 | activating transcription factor 1 |
|
atf1
|
ENSDARG00000109865 | activating transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf1 | dr11_v1_chr6_-_39521832_39521832 | 0.14 | 8.2e-01 | Click! |
Activity profile of atf1 motif
Sorted Z-values of atf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_12385308 | 2.05 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr6_+_27667359 | 1.88 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr15_-_163586 | 1.65 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr5_+_3501859 | 1.62 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr24_-_7632187 | 1.57 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr9_-_296169 | 1.54 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr23_-_4915118 | 1.47 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr23_+_28731379 | 1.34 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr16_+_36906693 | 1.34 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr23_-_29667716 | 1.34 |
ENSDART00000158302
ENSDART00000133902 |
clstn1
|
calsyntenin 1 |
| chr14_-_7888748 | 1.31 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr1_-_56223913 | 1.31 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr4_+_9669717 | 1.30 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr4_-_12388535 | 1.30 |
ENSDART00000017180
|
rergla
|
RERG/RAS-like a |
| chr16_-_29437373 | 1.30 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr20_+_34913069 | 1.28 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr24_-_6158933 | 1.22 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr2_-_11512819 | 1.21 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr2_-_42415902 | 1.21 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr19_+_24882845 | 1.18 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr1_-_38756870 | 1.16 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr21_-_43606502 | 1.14 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr23_-_29667544 | 1.13 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr3_-_49566364 | 1.13 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr3_+_15907297 | 1.11 |
ENSDART00000139206
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr8_+_44714336 | 1.11 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr9_+_219124 | 1.07 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr12_-_19103490 | 1.06 |
ENSDART00000060561
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr25_-_31863374 | 1.05 |
ENSDART00000028338
|
scamp5a
|
secretory carrier membrane protein 5a |
| chr21_-_23308286 | 1.04 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr5_+_37056818 | 1.04 |
ENSDART00000036760
|
tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr7_+_29133321 | 1.03 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr11_-_44163164 | 1.01 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr7_-_12065668 | 1.00 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr3_-_2593859 | 1.00 |
ENSDART00000143826
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr7_+_40228422 | 0.99 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr25_-_23052707 | 0.98 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr10_-_22845485 | 0.98 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr25_+_35683956 | 0.97 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr23_+_23183449 | 0.97 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr20_+_29743904 | 0.97 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr17_-_20979077 | 0.97 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr7_+_13382852 | 0.96 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr6_+_24817852 | 0.96 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr19_+_16222618 | 0.96 |
ENSDART00000137189
ENSDART00000169246 ENSDART00000190583 ENSDART00000189521 |
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr19_-_1961024 | 0.95 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr20_+_20637866 | 0.94 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr7_+_39166460 | 0.94 |
ENSDART00000052318
ENSDART00000146635 ENSDART00000173877 ENSDART00000173767 ENSDART00000173600 |
mdka
|
midkine a |
| chr15_-_27710513 | 0.93 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr10_-_36591511 | 0.92 |
ENSDART00000063347
|
slc1a3b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3b |
| chr5_-_45876908 | 0.91 |
ENSDART00000188229
|
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr18_+_21408794 | 0.90 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr3_-_1204341 | 0.90 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr10_+_32683089 | 0.89 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr20_+_20638034 | 0.88 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr9_-_54840124 | 0.88 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr12_-_30443562 | 0.88 |
ENSDART00000020769
|
adrb1
|
adrenoceptor beta 1 |
| chr17_-_26911852 | 0.86 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr17_+_8184649 | 0.85 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr7_+_26224211 | 0.84 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr2_+_20332044 | 0.84 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr13_-_31470439 | 0.83 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr21_+_11684830 | 0.82 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr16_+_34523515 | 0.81 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr5_-_63109232 | 0.81 |
ENSDART00000115128
|
usp2b
|
ubiquitin specific peptidase 2b |
| chr5_-_45877387 | 0.81 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr1_+_41666611 | 0.81 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr11_-_13341483 | 0.80 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr17_+_29345606 | 0.80 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr19_-_43552252 | 0.79 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr7_-_23563092 | 0.79 |
ENSDART00000132275
|
gpr185b
|
G protein-coupled receptor 185 b |
| chr13_+_23843712 | 0.78 |
ENSDART00000057611
|
oprm1
|
opioid receptor, mu 1 |
| chr10_-_24371312 | 0.78 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr21_-_23307653 | 0.77 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr4_-_16124417 | 0.77 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr15_+_28685625 | 0.77 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr21_+_11685009 | 0.77 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr11_+_40649412 | 0.77 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr18_-_19405616 | 0.77 |
ENSDART00000191290
ENSDART00000090855 |
megf11
|
multiple EGF-like-domains 11 |
| chr15_+_28685892 | 0.76 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr8_-_17771755 | 0.76 |
ENSDART00000063592
|
prkcz
|
protein kinase C, zeta |
| chr6_-_31348999 | 0.76 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr4_-_5764255 | 0.74 |
ENSDART00000113864
|
faxca
|
failed axon connections homolog a |
| chr6_-_58828398 | 0.74 |
ENSDART00000090634
|
kif5ab
|
kinesin family member 5A, b |
| chr17_-_11329959 | 0.74 |
ENSDART00000015418
|
irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr20_+_41549200 | 0.73 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr18_-_14743659 | 0.73 |
ENSDART00000125979
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr1_+_9290103 | 0.73 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr14_-_2270973 | 0.72 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr20_+_38724575 | 0.72 |
ENSDART00000015095
ENSDART00000152972 |
uts1
|
urotensin 1 |
| chr2_-_6292510 | 0.71 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr13_-_50672341 | 0.71 |
ENSDART00000190498
|
FP102122.1
|
|
| chr8_+_29742237 | 0.71 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr7_+_67486807 | 0.71 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr8_-_6943155 | 0.71 |
ENSDART00000139545
ENSDART00000033294 |
wdr13
|
WD repeat domain 13 |
| chr13_+_24834199 | 0.71 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr13_-_40499296 | 0.70 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr12_-_41619257 | 0.70 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr2_-_56635744 | 0.70 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr9_+_6997861 | 0.70 |
ENSDART00000190491
|
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr10_-_7858553 | 0.70 |
ENSDART00000182010
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr8_-_17067364 | 0.70 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr16_-_32649929 | 0.69 |
ENSDART00000136161
|
faxcb
|
failed axon connections homolog b |
| chr14_-_33454595 | 0.69 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr21_-_14878220 | 0.69 |
ENSDART00000131237
|
ulk1b
|
unc-51 like autophagy activating kinase 1 |
| chr19_+_10396042 | 0.69 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr8_-_54223316 | 0.68 |
ENSDART00000018054
|
trh
|
thyrotropin-releasing hormone |
| chr24_-_21172122 | 0.68 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr13_-_16191674 | 0.68 |
ENSDART00000147868
|
vwc2
|
von Willebrand factor C domain containing 2 |
| chr2_+_22694382 | 0.68 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr20_+_3108597 | 0.68 |
ENSDART00000133435
|
CEP170B (1 of many)
|
si:ch73-212j7.1 |
| chr1_-_22861348 | 0.68 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr10_+_23022263 | 0.67 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr2_+_20331445 | 0.67 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr19_-_32888758 | 0.67 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr9_+_6998082 | 0.67 |
ENSDART00000092480
ENSDART00000135576 ENSDART00000188884 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr25_-_31763897 | 0.66 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr8_-_39984593 | 0.66 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr16_-_9869056 | 0.66 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr5_-_23999777 | 0.66 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr16_-_17162843 | 0.65 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr3_-_18373604 | 0.65 |
ENSDART00000148132
|
spag9a
|
sperm associated antigen 9a |
| chr16_+_5184402 | 0.65 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr12_+_27243059 | 0.64 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr5_-_41560874 | 0.64 |
ENSDART00000136702
|
dnajb5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr23_+_45584223 | 0.64 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr1_-_19215336 | 0.63 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr19_+_45970692 | 0.63 |
ENSDART00000158781
|
si:ch211-153f2.7
|
si:ch211-153f2.7 |
| chr23_+_2136344 | 0.63 |
ENSDART00000085290
ENSDART00000092757 |
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr11_+_6819050 | 0.63 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr8_+_21588067 | 0.62 |
ENSDART00000172190
|
ajap1
|
adherens junctions associated protein 1 |
| chr4_+_5156117 | 0.62 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr3_-_18373425 | 0.61 |
ENSDART00000178522
|
spag9a
|
sperm associated antigen 9a |
| chr13_+_28705143 | 0.60 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr21_+_9576176 | 0.60 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr20_+_41549735 | 0.60 |
ENSDART00000184235
|
fam184a
|
family with sequence similarity 184, member A |
| chr3_-_62380146 | 0.60 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr1_-_54947592 | 0.60 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr5_-_24000211 | 0.59 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr11_-_6970107 | 0.59 |
ENSDART00000171255
|
COMP
|
si:ch211-43f4.1 |
| chr10_-_45379831 | 0.58 |
ENSDART00000186205
|
CABZ01117348.1
|
|
| chr17_-_15382704 | 0.58 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr15_-_28200049 | 0.58 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr24_-_6375774 | 0.58 |
ENSDART00000013294
|
anxa13
|
annexin A13 |
| chr18_-_19103929 | 0.58 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr7_-_44963154 | 0.57 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr3_+_61221194 | 0.57 |
ENSDART00000110030
|
lmtk2
|
lemur tyrosine kinase 2 |
| chr9_+_22631672 | 0.57 |
ENSDART00000101770
ENSDART00000126015 ENSDART00000145005 |
etv5a
|
ets variant 5a |
| chr5_-_57879138 | 0.56 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr1_+_45323400 | 0.56 |
ENSDART00000148906
ENSDART00000132366 |
emp1
|
epithelial membrane protein 1 |
| chr22_-_21392748 | 0.56 |
ENSDART00000144648
|
ankrd24
|
ankyrin repeat domain 24 |
| chr20_-_1191910 | 0.56 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr10_+_44903676 | 0.56 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr7_+_7552008 | 0.55 |
ENSDART00000173018
ENSDART00000049311 |
clcn3
|
chloride channel 3 |
| chr2_-_58201173 | 0.55 |
ENSDART00000166282
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr16_+_26766423 | 0.55 |
ENSDART00000048036
|
gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr17_+_23554932 | 0.54 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr5_-_20814576 | 0.54 |
ENSDART00000098682
ENSDART00000147639 |
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr20_+_36682051 | 0.54 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr8_+_3085120 | 0.54 |
ENSDART00000148020
ENSDART00000136250 |
lhx2b
|
LIM homeobox 2b |
| chr23_-_33654889 | 0.53 |
ENSDART00000146180
|
csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr1_+_10305611 | 0.53 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr14_+_790166 | 0.53 |
ENSDART00000123912
|
adra2da
|
adrenergic, alpha-2D-, receptor a |
| chr25_-_24074500 | 0.53 |
ENSDART00000040410
|
th
|
tyrosine hydroxylase |
| chr12_+_32159272 | 0.52 |
ENSDART00000153167
|
hlfb
|
hepatic leukemia factor b |
| chr22_-_600016 | 0.52 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr7_-_48805181 | 0.52 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr19_-_10395683 | 0.52 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr2_-_30324610 | 0.52 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr2_+_35603637 | 0.52 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr15_+_5116179 | 0.52 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr4_-_69189894 | 0.51 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr3_-_62393449 | 0.51 |
ENSDART00000101870
ENSDART00000140782 ENSDART00000181704 |
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr5_+_62374092 | 0.51 |
ENSDART00000082965
|
CU928117.1
|
|
| chr21_+_25054420 | 0.51 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr20_+_2950005 | 0.50 |
ENSDART00000135919
|
amd1
|
adenosylmethionine decarboxylase 1 |
| chr14_+_8947282 | 0.50 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr9_-_13963078 | 0.50 |
ENSDART00000193398
ENSDART00000061156 |
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr10_+_37145007 | 0.50 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr18_-_5781922 | 0.50 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr4_+_74131530 | 0.50 |
ENSDART00000174125
|
CABZ01072523.1
|
|
| chr6_-_46403475 | 0.50 |
ENSDART00000154148
|
camk1a
|
calcium/calmodulin-dependent protein kinase Ia |
| chr13_-_3155243 | 0.49 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr11_-_18800299 | 0.49 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr21_-_25522510 | 0.49 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr13_-_40754499 | 0.49 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr2_+_11685742 | 0.49 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr19_-_3488860 | 0.48 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr4_-_1801519 | 0.48 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr1_-_11291324 | 0.48 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr6_-_25952848 | 0.48 |
ENSDART00000076997
ENSDART00000148748 |
lmo4b
|
LIM domain only 4b |
| chr14_-_48939560 | 0.48 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr9_+_42546504 | 0.47 |
ENSDART00000192224
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr9_-_31524907 | 0.47 |
ENSDART00000142904
ENSDART00000127214 ENSDART00000133427 ENSDART00000146268 ENSDART00000182541 ENSDART00000184736 |
tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr8_+_50983551 | 0.47 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr4_+_73973242 | 0.47 |
ENSDART00000182529
|
LO018181.1
|
|
| chr16_+_25137483 | 0.47 |
ENSDART00000155666
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr12_-_41618844 | 0.47 |
ENSDART00000160054
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr16_-_45069882 | 0.47 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr12_+_28298859 | 0.46 |
ENSDART00000183366
|
LO017791.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of atf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 1.6 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.4 | 1.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 3.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.3 | 0.9 | GO:0097378 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 1.0 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.2 | 1.1 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 0.6 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.2 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 0.6 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 1.0 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 1.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.2 | 0.7 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 0.5 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.2 | 0.9 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.2 | 2.0 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.2 | 0.8 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.2 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 2.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.5 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.9 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.8 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.4 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.4 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 2.3 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.2 | GO:1990592 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.5 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 1.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.4 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.5 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.4 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.1 | 0.7 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 1.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.7 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 0.5 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 1.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.1 | 0.5 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.0 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.3 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 2.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 1.9 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.2 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.2 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.1 | 0.3 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.4 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.1 | 0.8 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.1 | 0.4 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 1.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.9 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.9 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0014005 | microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 1.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.6 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.3 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.0 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.4 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.1 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0032262 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 1.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.3 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 1.0 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.0 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.0 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.0 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 4.3 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.1 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.0 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.1 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.0 | 0.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0090311 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.0 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.0 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.3 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.6 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.4 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.0 | 0.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.1 | GO:0050771 | negative regulation of axon extension(GO:0030517) negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 2.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 1.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 1.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.6 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.4 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 4.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 1.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 3.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 3.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 1.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 0.8 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.2 | 1.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 0.9 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.2 | 1.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.6 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 1.9 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.4 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 1.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.4 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 1.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 0.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 4.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.5 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 3.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.5 | GO:0051379 | alpha2-adrenergic receptor activity(GO:0004938) epinephrine binding(GO:0051379) |
| 0.1 | 0.7 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 1.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 2.0 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.6 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.6 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.3 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 1.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 5.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.4 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 1.0 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 3.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.7 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.0 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 1.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |