Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
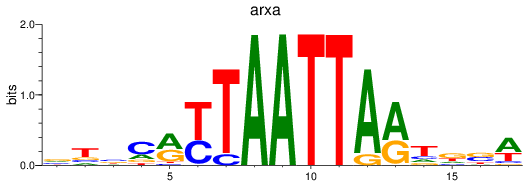
Results for arxa
Z-value: 1.25
Transcription factors associated with arxa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arxa
|
ENSDARG00000058011 | aristaless related homeobox a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arxa | dr11_v1_chr24_-_23942722_23942722 | -0.84 | 7.8e-02 | Click! |
Activity profile of arxa motif
Sorted Z-values of arxa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_11775269 | 1.83 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr3_-_59297532 | 1.26 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr20_-_37813863 | 1.22 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr4_-_9891874 | 1.18 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr9_-_9415000 | 1.09 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr12_-_35830625 | 0.91 |
ENSDART00000180028
|
CU459056.1
|
|
| chr25_-_21031007 | 0.81 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr12_+_22580579 | 0.78 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr11_+_705727 | 0.78 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr14_+_901847 | 0.76 |
ENSDART00000166991
|
si:ch73-208h1.2
|
si:ch73-208h1.2 |
| chr2_-_36918709 | 0.74 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr23_-_33350990 | 0.73 |
ENSDART00000144831
|
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr19_+_43780970 | 0.72 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr25_-_28674739 | 0.70 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr14_+_34558480 | 0.67 |
ENSDART00000075170
|
pttg1
|
pituitary tumor-transforming 1 |
| chr23_-_30041065 | 0.66 |
ENSDART00000131209
ENSDART00000127192 |
ccdc187
|
coiled-coil domain containing 187 |
| chr20_+_11731039 | 0.64 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr6_-_7720332 | 0.64 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr5_-_7199998 | 0.63 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr23_-_16485190 | 0.62 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr1_+_44127292 | 0.60 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr16_-_11798994 | 0.60 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr6_+_36839509 | 0.60 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr19_+_2631565 | 0.59 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr7_-_50410524 | 0.56 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr23_+_4709607 | 0.56 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr13_+_24756486 | 0.54 |
ENSDART00000137074
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr13_-_31017960 | 0.53 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr2_-_26596794 | 0.52 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr14_-_14659023 | 0.50 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr3_-_29941357 | 0.50 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr20_-_43663494 | 0.49 |
ENSDART00000144564
|
BX470188.1
|
|
| chr9_-_32656656 | 0.49 |
ENSDART00000078399
|
gzm3
|
granzyme 3, tandem duplicate 1 |
| chr6_+_39905021 | 0.49 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr12_-_48188928 | 0.48 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr25_-_13319112 | 0.48 |
ENSDART00000179885
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr22_+_19366866 | 0.47 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr24_-_36680261 | 0.47 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr8_+_26059677 | 0.46 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr16_-_51271962 | 0.46 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr13_-_36798204 | 0.46 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr17_+_20589553 | 0.46 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr10_+_11261576 | 0.46 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr20_+_25586099 | 0.45 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr14_-_4145594 | 0.44 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr8_+_28695914 | 0.44 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr12_-_36521767 | 0.43 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr5_-_42083363 | 0.43 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr21_+_43328685 | 0.43 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr23_-_37575030 | 0.42 |
ENSDART00000031875
|
tor1l3
|
torsin family 1 like 3 |
| chr16_+_23799622 | 0.42 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr5_-_42082730 | 0.42 |
ENSDART00000135625
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr18_+_13275735 | 0.42 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr23_-_42232124 | 0.41 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr23_+_26733232 | 0.41 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr7_+_21272833 | 0.41 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr24_-_37640705 | 0.40 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr6_-_55399214 | 0.39 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr15_-_4528326 | 0.39 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr25_+_11008419 | 0.38 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr7_+_24573721 | 0.38 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr5_+_71802014 | 0.38 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr24_+_19415124 | 0.38 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr13_-_2010191 | 0.38 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr20_+_19176959 | 0.38 |
ENSDART00000163276
ENSDART00000020099 |
blk
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr8_+_25079470 | 0.36 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr15_-_38129845 | 0.36 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr20_+_25225112 | 0.36 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr3_+_27798094 | 0.35 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr9_+_24065855 | 0.35 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr21_-_44081540 | 0.34 |
ENSDART00000130833
|
FO704810.1
|
|
| chr24_+_29912509 | 0.33 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr20_+_38458084 | 0.32 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr19_+_42955280 | 0.32 |
ENSDART00000141225
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr22_+_7738966 | 0.32 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr18_-_46258612 | 0.32 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr6_-_40842768 | 0.31 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr7_+_19495905 | 0.31 |
ENSDART00000125584
ENSDART00000173774 |
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr23_+_1029450 | 0.31 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr2_-_10877765 | 0.31 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr13_+_646700 | 0.30 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr23_+_36460239 | 0.30 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr7_+_22543963 | 0.29 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr13_-_37229245 | 0.29 |
ENSDART00000140923
|
si:dkeyp-77c8.5
|
si:dkeyp-77c8.5 |
| chr15_-_35134918 | 0.28 |
ENSDART00000157196
|
si:ch211-272b8.7
|
si:ch211-272b8.7 |
| chr18_+_39106722 | 0.28 |
ENSDART00000122377
|
bcl2l10
|
BCL2 like 10 |
| chr14_-_413273 | 0.28 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr1_-_17715493 | 0.28 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr20_-_51355465 | 0.27 |
ENSDART00000151620
ENSDART00000151690 ENSDART00000110289 |
tcte1
|
t-complex-associated-testis-expressed 1 |
| chr14_+_40874608 | 0.27 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr21_-_19314618 | 0.27 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr22_-_5529241 | 0.27 |
ENSDART00000157893
|
CABZ01064968.1
|
|
| chr15_+_41932069 | 0.27 |
ENSDART00000143007
|
si:ch211-191a16.2
|
si:ch211-191a16.2 |
| chr5_-_4418555 | 0.26 |
ENSDART00000170158
|
apooa
|
apolipoprotein O, a |
| chr1_+_51721851 | 0.26 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr7_+_26173751 | 0.26 |
ENSDART00000065131
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr19_+_12237945 | 0.26 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr21_-_45363871 | 0.26 |
ENSDART00000075443
ENSDART00000182078 ENSDART00000151106 |
PPP2CA
|
zgc:56064 |
| chr3_-_19368435 | 0.25 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr12_-_28363111 | 0.25 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr12_-_4185554 | 0.25 |
ENSDART00000152313
|
si:dkey-32n7.7
|
si:dkey-32n7.7 |
| chr2_-_38284648 | 0.25 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr24_-_29586082 | 0.24 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr10_-_3138859 | 0.24 |
ENSDART00000190606
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr24_+_37640626 | 0.24 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr21_+_45223194 | 0.24 |
ENSDART00000150902
|
si:ch73-269m14.3
|
si:ch73-269m14.3 |
| chr25_+_22320738 | 0.24 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr10_-_3138403 | 0.24 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr22_+_737211 | 0.24 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr25_+_30131055 | 0.23 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr16_+_23984179 | 0.23 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr1_+_47499888 | 0.23 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr10_-_2788668 | 0.23 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr4_+_20566371 | 0.22 |
ENSDART00000127576
|
BX248410.1
|
|
| chr7_+_2969661 | 0.22 |
ENSDART00000158382
|
CABZ01007829.1
|
|
| chr7_+_19495379 | 0.22 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr7_+_34453185 | 0.22 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr10_+_38610741 | 0.22 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr10_-_43771447 | 0.22 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr25_+_13620555 | 0.22 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr2_+_9990491 | 0.22 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr8_+_17167876 | 0.22 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr5_+_69747417 | 0.21 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr9_-_30555725 | 0.21 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr20_-_9095105 | 0.21 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr14_+_30568961 | 0.21 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr6_+_12527725 | 0.20 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr9_+_22351443 | 0.20 |
ENSDART00000080054
|
crygs3
|
crystallin, gamma S3 |
| chr16_-_38118003 | 0.20 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr24_-_38657683 | 0.20 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr1_-_45616470 | 0.20 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr23_+_11347313 | 0.20 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr22_+_19407531 | 0.20 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr4_-_67980261 | 0.20 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr11_-_39118882 | 0.20 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr6_+_8598428 | 0.20 |
ENSDART00000032118
|
ube2g2
|
ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) |
| chr2_-_9989919 | 0.19 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr2_-_59205393 | 0.19 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr3_-_58798377 | 0.19 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr3_+_30501135 | 0.19 |
ENSDART00000165869
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr1_+_35985813 | 0.19 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr16_+_19029297 | 0.18 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr1_+_47165842 | 0.18 |
ENSDART00000053152
ENSDART00000167051 |
cbr1
|
carbonyl reductase 1 |
| chr20_-_40360571 | 0.18 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr16_-_32013913 | 0.18 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr19_-_5669122 | 0.17 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr4_+_51551318 | 0.17 |
ENSDART00000190283
|
si:dkey-165e24.1
|
si:dkey-165e24.1 |
| chr20_+_18661624 | 0.17 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr3_+_32118670 | 0.17 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr9_+_43799829 | 0.17 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr4_-_9909371 | 0.17 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr5_-_16472719 | 0.17 |
ENSDART00000162071
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr15_+_25156382 | 0.17 |
ENSDART00000171092
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr8_+_49065348 | 0.17 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr1_-_54063520 | 0.17 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr18_+_7073130 | 0.17 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr23_-_19225709 | 0.16 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_+_1988036 | 0.16 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr11_-_16152400 | 0.16 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr4_+_5255041 | 0.16 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr11_+_31324335 | 0.16 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr13_+_7049823 | 0.16 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr3_+_29445473 | 0.16 |
ENSDART00000020381
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr24_-_7995960 | 0.16 |
ENSDART00000186594
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr3_-_34052882 | 0.16 |
ENSDART00000151463
|
ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr6_-_52788213 | 0.16 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr7_-_60156409 | 0.15 |
ENSDART00000006802
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr16_-_7793457 | 0.15 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr5_-_11809710 | 0.15 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr6_+_40922572 | 0.14 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr7_-_30677104 | 0.14 |
ENSDART00000188360
|
myo1ea
|
myosin IE, a |
| chr14_-_7045009 | 0.14 |
ENSDART00000112082
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr5_+_65871688 | 0.14 |
ENSDART00000175217
|
FAM163B (1 of many)
|
family with sequence similarity 163 member B |
| chr2_-_45331115 | 0.14 |
ENSDART00000158003
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr5_+_66433287 | 0.14 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr19_-_47323267 | 0.13 |
ENSDART00000190077
|
CU138512.1
|
|
| chr18_+_35128685 | 0.13 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr19_+_1688727 | 0.13 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr16_-_13818061 | 0.12 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr10_+_26652859 | 0.12 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
| chr5_+_66312359 | 0.12 |
ENSDART00000137534
|
malt1
|
MALT paracaspase 1 |
| chr14_+_30795559 | 0.12 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr17_+_50524573 | 0.12 |
ENSDART00000187974
|
CR382385.1
|
|
| chr13_+_35339182 | 0.12 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr21_-_30082414 | 0.12 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr9_-_34937025 | 0.12 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr20_-_5291012 | 0.11 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr11_-_41853874 | 0.11 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr4_+_59845617 | 0.11 |
ENSDART00000167626
ENSDART00000123157 |
si:dkey-196n19.2
|
si:dkey-196n19.2 |
| chr14_-_7137808 | 0.11 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr22_+_30137374 | 0.11 |
ENSDART00000187808
|
add3a
|
adducin 3 (gamma) a |
| chr4_-_69428110 | 0.11 |
ENSDART00000160607
|
si:ch211-145h19.3
|
si:ch211-145h19.3 |
| chr2_+_10878406 | 0.11 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr6_+_7533601 | 0.11 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr15_+_37965953 | 0.11 |
ENSDART00000154192
|
si:dkey-238d18.10
|
si:dkey-238d18.10 |
| chr1_-_57629639 | 0.10 |
ENSDART00000158984
|
zmp:0000001289
|
zmp:0000001289 |
| chr20_-_2949028 | 0.10 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr17_-_49978986 | 0.10 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr8_-_53044300 | 0.10 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr23_-_36303216 | 0.10 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of arxa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.4 | GO:0002902 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 0.4 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.7 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.4 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.5 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.7 | GO:0033048 | negative regulation of sister chromatid segregation(GO:0033046) negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.3 | GO:0010522 | regulation of calcium ion transport into cytosol(GO:0010522) regulation of release of sequestered calcium ion into cytosol(GO:0051279) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.9 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.9 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0010965 | regulation of mitotic sister chromatid separation(GO:0010965) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 0.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.2 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |