Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
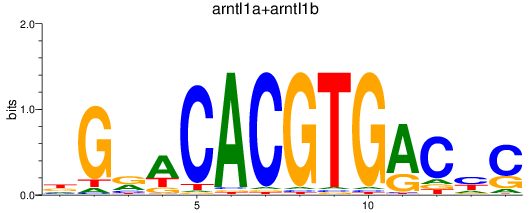
Results for arntl1a+arntl1b
Z-value: 1.16
Transcription factors associated with arntl1a+arntl1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arntl1a
|
ENSDARG00000006791 | aryl hydrocarbon receptor nuclear translocator-like 1a |
|
arntl1b
|
ENSDARG00000035732 | aryl hydrocarbon receptor nuclear translocator-like 1b |
|
arntl1b
|
ENSDARG00000114562 | aryl hydrocarbon receptor nuclear translocator-like 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arntl1b | dr11_v1_chr7_+_66048102_66048102 | 0.79 | 1.1e-01 | Click! |
| arntl1a | dr11_v1_chr25_-_17918536_17918536 | 0.37 | 5.4e-01 | Click! |
Activity profile of arntl1a+arntl1b motif
Sorted Z-values of arntl1a+arntl1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_13696537 | 1.60 |
ENSDART00000109195
ENSDART00000122041 ENSDART00000192554 |
nrcama
|
neuronal cell adhesion molecule a |
| chr7_+_48999723 | 1.52 |
ENSDART00000182699
ENSDART00000166329 |
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr24_-_7632187 | 1.21 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr10_-_6775271 | 1.19 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr24_-_33756003 | 0.91 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr13_+_421231 | 0.91 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr9_+_42095220 | 0.89 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr7_-_13381129 | 0.89 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr25_+_25123385 | 0.86 |
ENSDART00000163892
|
ldha
|
lactate dehydrogenase A4 |
| chr10_+_23022263 | 0.86 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr17_+_8183393 | 0.82 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr9_-_24413008 | 0.81 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr2_+_50626476 | 0.81 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr9_-_44295071 | 0.81 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr9_+_42066030 | 0.80 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr18_+_26337869 | 0.80 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr21_-_4539899 | 0.79 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr22_-_3595439 | 0.76 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr2_+_6181383 | 0.76 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr25_+_7423770 | 0.71 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr20_+_40766645 | 0.69 |
ENSDART00000144401
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr13_-_31441042 | 0.67 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr15_+_1148074 | 0.67 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr8_-_6943155 | 0.65 |
ENSDART00000139545
ENSDART00000033294 |
wdr13
|
WD repeat domain 13 |
| chr15_-_3094516 | 0.64 |
ENSDART00000179719
|
slitrk5a
|
SLIT and NTRK-like family, member 5a |
| chr8_+_41533268 | 0.63 |
ENSDART00000142377
|
si:ch211-158d24.2
|
si:ch211-158d24.2 |
| chr7_-_30082931 | 0.63 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr1_+_29178117 | 0.62 |
ENSDART00000114536
|
si:ch211-198k9.6
|
si:ch211-198k9.6 |
| chr18_+_17428258 | 0.61 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr7_-_40993456 | 0.56 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr21_+_31150438 | 0.55 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_+_17695426 | 0.55 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr23_-_16692312 | 0.55 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr8_+_35032633 | 0.54 |
ENSDART00000184683
ENSDART00000184109 |
zgc:77614
|
zgc:77614 |
| chr18_+_39074139 | 0.54 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr1_+_29178331 | 0.54 |
ENSDART00000186905
|
si:ch211-198k9.6
|
si:ch211-198k9.6 |
| chr14_+_25465346 | 0.53 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr6_-_39605734 | 0.53 |
ENSDART00000044276
ENSDART00000179059 |
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr7_-_41812015 | 0.52 |
ENSDART00000174058
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr12_-_18483348 | 0.51 |
ENSDART00000152757
|
tex2l
|
testis expressed 2, like |
| chr16_-_9869056 | 0.51 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr17_-_14705039 | 0.51 |
ENSDART00000154281
ENSDART00000123550 |
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr8_+_34988481 | 0.51 |
ENSDART00000186808
ENSDART00000164942 ENSDART00000186472 ENSDART00000182717 |
zgc:174461
|
zgc:174461 |
| chr22_-_11136625 | 0.51 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr20_-_40766387 | 0.50 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr7_-_49654492 | 0.50 |
ENSDART00000174324
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr21_-_42100471 | 0.49 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr19_-_5103313 | 0.48 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr14_+_2243 | 0.48 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr8_+_34434345 | 0.48 |
ENSDART00000190246
ENSDART00000189447 ENSDART00000185557 ENSDART00000189230 |
zgc:174461
|
zgc:174461 |
| chr8_+_34476532 | 0.47 |
ENSDART00000181956
ENSDART00000191537 |
zgc:174461
|
zgc:174461 |
| chr8_+_34998570 | 0.47 |
ENSDART00000184294
|
zgc:77614
|
zgc:77614 |
| chr18_-_19103929 | 0.47 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr18_+_31280984 | 0.47 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr8_+_34443759 | 0.46 |
ENSDART00000187788
ENSDART00000187679 ENSDART00000188872 ENSDART00000186358 |
zgc:174461
|
zgc:174461 |
| chr8_+_34439052 | 0.46 |
ENSDART00000193021
ENSDART00000193851 ENSDART00000183170 ENSDART00000181227 |
zgc:174461
|
zgc:174461 |
| chr3_+_62353650 | 0.44 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr22_+_38159823 | 0.44 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr11_-_43473824 | 0.44 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr8_+_34448496 | 0.44 |
ENSDART00000188072
ENSDART00000180224 ENSDART00000189510 ENSDART00000190390 |
zgc:174461
|
zgc:174461 |
| chr12_-_17424162 | 0.43 |
ENSDART00000079144
ENSDART00000079138 |
ptenb
|
phosphatase and tensin homolog B |
| chr7_+_34620418 | 0.43 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr8_+_34567717 | 0.43 |
ENSDART00000141506
|
BX005399.1
|
|
| chr8_+_34531954 | 0.43 |
ENSDART00000184878
ENSDART00000192417 ENSDART00000184988 ENSDART00000192039 |
zgc:174461
|
zgc:174461 |
| chr8_+_34540801 | 0.43 |
ENSDART00000186390
ENSDART00000184590 ENSDART00000187379 ENSDART00000180922 ENSDART00000187547 |
zgc:174461
|
zgc:174461 |
| chr8_+_34562983 | 0.43 |
ENSDART00000179915
ENSDART00000182870 ENSDART00000191069 ENSDART00000193201 ENSDART00000187830 |
zgc:174461
|
zgc:174461 |
| chr8_+_34927665 | 0.43 |
ENSDART00000186136
ENSDART00000192243 ENSDART00000182256 ENSDART00000186057 |
zgc:174461
|
zgc:174461 |
| chr8_+_34685407 | 0.43 |
ENSDART00000111653
ENSDART00000186260 ENSDART00000190456 ENSDART00000184348 ENSDART00000180741 |
zgc:174461
|
zgc:174461 |
| chr9_-_27805801 | 0.43 |
ENSDART00000140608
ENSDART00000114542 |
si:rp71-45g20.10
|
si:rp71-45g20.10 |
| chr8_+_34486656 | 0.43 |
ENSDART00000136887
ENSDART00000160081 ENSDART00000162349 ENSDART00000183007 ENSDART00000185735 |
zgc:77614
|
zgc:77614 |
| chr8_+_34572359 | 0.43 |
ENSDART00000187498
ENSDART00000184035 ENSDART00000193944 ENSDART00000185472 ENSDART00000193433 |
zgc:174461
|
zgc:174461 |
| chr2_-_33687214 | 0.43 |
ENSDART00000147439
|
atp6v0b
|
ATPase H+ transporting V0 subunit b |
| chr8_+_34527239 | 0.42 |
ENSDART00000190604
ENSDART00000179883 ENSDART00000180594 ENSDART00000183138 ENSDART00000183360 |
zgc:174461
|
zgc:174461 |
| chr8_+_34932397 | 0.41 |
ENSDART00000193631
ENSDART00000188761 ENSDART00000181280 ENSDART00000191784 ENSDART00000187029 |
zgc:174461
|
zgc:174461 |
| chr2_-_32486080 | 0.40 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr8_+_34720244 | 0.39 |
ENSDART00000181958
ENSDART00000189806 ENSDART00000190167 ENSDART00000183165 |
zgc:174461
|
zgc:174461 |
| chr8_+_34922981 | 0.39 |
ENSDART00000188445
ENSDART00000188818 ENSDART00000182965 ENSDART00000180718 |
BX005361.1
|
|
| chr22_-_1237003 | 0.39 |
ENSDART00000169746
|
ampd2a
|
adenosine monophosphate deaminase 2a |
| chr9_-_18742704 | 0.39 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr18_+_6479963 | 0.38 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr23_-_10786400 | 0.38 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr8_+_34629184 | 0.37 |
ENSDART00000188917
ENSDART00000180349 ENSDART00000191426 |
zgc:174461
|
zgc:174461 |
| chr13_-_40120252 | 0.37 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr3_-_25421504 | 0.37 |
ENSDART00000154200
|
bptf
|
bromodomain PHD finger transcription factor |
| chr20_-_2355357 | 0.37 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr22_-_29083070 | 0.36 |
ENSDART00000104812
ENSDART00000172576 |
cbx6a
|
chromobox homolog 6a |
| chr1_+_6189949 | 0.36 |
ENSDART00000092277
|
retreg2
|
reticulophagy regulator family member 2 |
| chr18_+_17428506 | 0.36 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr7_+_1442059 | 0.36 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr8_+_21146262 | 0.35 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr25_+_17589906 | 0.35 |
ENSDART00000167750
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr25_+_37480285 | 0.35 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr6_+_54498220 | 0.35 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr15_-_1534232 | 0.34 |
ENSDART00000056763
ENSDART00000133943 |
ift80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr11_+_44300548 | 0.34 |
ENSDART00000191626
|
CABZ01100304.1
|
|
| chr3_-_9722603 | 0.34 |
ENSDART00000168234
|
crebbpb
|
CREB binding protein b |
| chr10_-_41157135 | 0.33 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr11_+_14199802 | 0.33 |
ENSDART00000102520
ENSDART00000133172 |
palm1a
|
paralemmin 1a |
| chr25_+_37268900 | 0.33 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr22_+_15979430 | 0.33 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr4_+_331351 | 0.33 |
ENSDART00000132625
|
tulp4a
|
tubby like protein 4a |
| chr16_+_13822137 | 0.33 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr13_+_19884631 | 0.33 |
ENSDART00000089533
|
atrnl1a
|
attractin-like 1a |
| chr10_-_1320114 | 0.33 |
ENSDART00000073617
|
opn4xa
|
opsin 4xa |
| chr5_-_55395964 | 0.32 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr24_+_32472155 | 0.32 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr4_+_13640903 | 0.32 |
ENSDART00000155349
|
pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr6_-_36552844 | 0.31 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr18_-_6881392 | 0.31 |
ENSDART00000154968
|
si:dkey-266m15.7
|
si:dkey-266m15.7 |
| chr1_+_32013688 | 0.31 |
ENSDART00000168045
|
BX901898.2
|
|
| chr10_+_22891126 | 0.30 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr7_-_41812355 | 0.30 |
ENSDART00000016105
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr6_-_13709591 | 0.29 |
ENSDART00000151771
|
chpfb
|
chondroitin polymerizing factor b |
| chr3_-_25420931 | 0.29 |
ENSDART00000109601
ENSDART00000182184 |
bptf
|
bromodomain PHD finger transcription factor |
| chr3_+_39568290 | 0.29 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr9_+_33788626 | 0.29 |
ENSDART00000088469
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr2_-_51096647 | 0.29 |
ENSDART00000167172
|
THEM6
|
si:ch73-52e5.2 |
| chr4_+_842010 | 0.28 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr19_-_38830582 | 0.28 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr22_+_30184039 | 0.28 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr5_-_37881345 | 0.28 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr13_-_9841806 | 0.28 |
ENSDART00000101949
|
sfxn4
|
sideroflexin 4 |
| chr3_+_24511959 | 0.28 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr13_-_3474373 | 0.28 |
ENSDART00000157437
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr9_-_3149896 | 0.27 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr11_-_43104475 | 0.27 |
ENSDART00000125368
|
acyp2
|
acylphosphatase 2, muscle type |
| chr20_+_32118559 | 0.27 |
ENSDART00000026273
|
cd164
|
CD164 molecule, sialomucin |
| chr21_-_14251306 | 0.27 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr16_+_53387085 | 0.27 |
ENSDART00000154223
ENSDART00000101404 |
kif13a
|
kinesin family member 13A |
| chr24_-_14711597 | 0.26 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr20_-_32270866 | 0.26 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr24_+_3307857 | 0.26 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr25_+_17590095 | 0.26 |
ENSDART00000009767
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr9_-_18743012 | 0.25 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr16_+_7985886 | 0.24 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr24_-_13364311 | 0.24 |
ENSDART00000183808
|
KCNB2
|
si:dkey-192j17.1 |
| chr18_+_8231138 | 0.24 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr14_+_28442963 | 0.23 |
ENSDART00000186495
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr22_-_38360205 | 0.23 |
ENSDART00000162055
|
mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr8_+_34624025 | 0.23 |
ENSDART00000180772
|
BX247865.2
|
|
| chr18_+_19990412 | 0.23 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr11_+_29770966 | 0.23 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr19_-_43552252 | 0.23 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr12_-_26587231 | 0.23 |
ENSDART00000153168
|
slc16a5a
|
solute carrier family 16 (monocarboxylate transporter), member 5a |
| chr4_-_13921185 | 0.22 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr7_+_55633483 | 0.22 |
ENSDART00000180993
ENSDART00000184845 |
trappc2l
|
trafficking protein particle complex 2-like |
| chr20_+_50115335 | 0.22 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr5_+_15992655 | 0.22 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr8_+_19674369 | 0.22 |
ENSDART00000138176
|
foxd2
|
forkhead box D2 |
| chr12_-_7280551 | 0.21 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr12_-_34827477 | 0.21 |
ENSDART00000153026
|
NDUFAF8
|
si:dkey-21c1.6 |
| chr12_+_13652361 | 0.21 |
ENSDART00000182757
ENSDART00000152689 |
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr12_+_34827808 | 0.21 |
ENSDART00000105533
|
tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr17_+_30448452 | 0.21 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr8_-_34767412 | 0.20 |
ENSDART00000164901
|
CABZ01049825.1
|
|
| chr19_-_5103141 | 0.20 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_+_15817644 | 0.20 |
ENSDART00000055787
|
natd1
|
zgc:110779 |
| chr24_+_35911300 | 0.20 |
ENSDART00000129679
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr19_+_40379771 | 0.20 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr3_-_16055432 | 0.20 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr10_+_22890791 | 0.19 |
ENSDART00000176011
|
arrb2a
|
arrestin, beta 2a |
| chr1_-_28473350 | 0.19 |
ENSDART00000190608
ENSDART00000148175 |
si:ch1073-440b2.1
|
si:ch1073-440b2.1 |
| chr21_+_43178831 | 0.19 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr1_+_50639416 | 0.19 |
ENSDART00000141977
|
herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr4_-_2036620 | 0.19 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr20_-_33174899 | 0.19 |
ENSDART00000047834
|
nbas
|
neuroblastoma amplified sequence |
| chr23_+_2825940 | 0.19 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr12_+_13652747 | 0.18 |
ENSDART00000066359
|
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr20_-_51814080 | 0.18 |
ENSDART00000041476
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr16_+_14216581 | 0.18 |
ENSDART00000113093
|
gba
|
glucosidase, beta, acid |
| chr24_+_35911020 | 0.18 |
ENSDART00000088480
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr21_+_11415224 | 0.18 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr9_-_27805644 | 0.18 |
ENSDART00000192431
|
si:rp71-45g20.10
|
si:rp71-45g20.10 |
| chr1_+_56447107 | 0.18 |
ENSDART00000091924
|
CABZ01059392.1
|
|
| chr11_+_5499661 | 0.18 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr25_-_3469576 | 0.18 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr7_+_7048245 | 0.17 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr24_+_35183595 | 0.17 |
ENSDART00000075142
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr20_+_13969414 | 0.17 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr15_+_1199407 | 0.17 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr7_-_72426484 | 0.17 |
ENSDART00000190063
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr4_+_8670662 | 0.17 |
ENSDART00000168768
|
adipor2
|
adiponectin receptor 2 |
| chr16_+_22865942 | 0.16 |
ENSDART00000103235
ENSDART00000143957 |
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr1_-_54765262 | 0.16 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr15_-_44077937 | 0.16 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr21_+_6780340 | 0.16 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr9_-_27771182 | 0.16 |
ENSDART00000170931
|
si:rp71-45g20.11
|
si:rp71-45g20.11 |
| chr9_+_54686686 | 0.16 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr13_+_281214 | 0.16 |
ENSDART00000137572
|
mpc1
|
mitochondrial pyruvate carrier 1 |
| chr17_+_43629008 | 0.15 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr7_-_50272912 | 0.15 |
ENSDART00000098842
|
hsd17b12b
|
hydroxysteroid (17-beta) dehydrogenase 12b |
| chr9_+_33788389 | 0.15 |
ENSDART00000144623
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr17_+_50074372 | 0.15 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr10_-_35220285 | 0.14 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr9_-_27771339 | 0.14 |
ENSDART00000135722
ENSDART00000140381 |
si:rp71-45g20.11
|
si:rp71-45g20.11 |
| chr3_-_1190132 | 0.14 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr4_+_16725960 | 0.14 |
ENSDART00000034441
|
tcp11l2
|
t-complex 11, testis-specific-like 2 |
| chr21_-_36948 | 0.14 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr5_-_43959972 | 0.14 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr23_-_46034609 | 0.14 |
ENSDART00000158712
|
zgc:65873
|
zgc:65873 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arntl1a+arntl1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.7 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.4 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 0.2 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 0.2 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.2 | GO:0072388 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.8 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.2 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.7 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.8 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.4 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.3 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 1.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.9 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.6 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.1 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.7 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 0.5 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.2 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.0 | 0.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.7 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.0 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |