Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
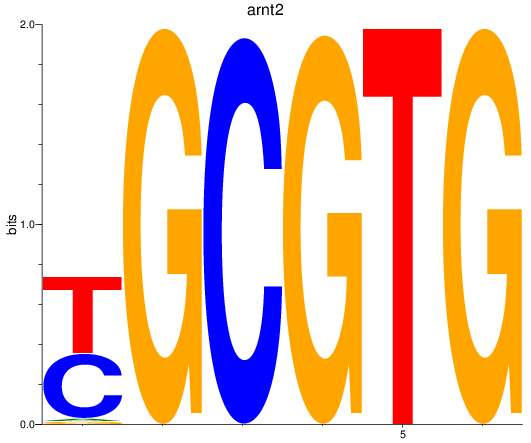
Results for arnt2
Z-value: 5.62
Transcription factors associated with arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arnt2
|
ENSDARG00000103697 | aryl-hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arnt2 | dr11_v1_chr7_+_10701938_10701938 | 0.95 | 1.4e-02 | Click! |
Activity profile of arnt2 motif
Sorted Z-values of arnt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_23429228 | 6.74 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr20_+_35382482 | 6.52 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr9_-_44295071 | 5.70 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr18_+_7345417 | 5.05 |
ENSDART00000041429
|
glipr1b
|
GLI pathogenesis-related 1b |
| chr13_+_27314795 | 4.82 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr11_+_7324704 | 4.61 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr5_+_32162684 | 4.59 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr5_+_36415978 | 4.41 |
ENSDART00000084464
|
fam155b
|
family with sequence similarity 155, member B |
| chr12_+_35119762 | 4.21 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr21_+_5169154 | 4.14 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr4_-_4780667 | 3.92 |
ENSDART00000133973
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr5_+_55626693 | 3.84 |
ENSDART00000168908
ENSDART00000161412 |
ntrk2b
|
neurotrophic tyrosine kinase, receptor, type 2b |
| chr5_+_23118470 | 3.78 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr24_-_3419998 | 3.67 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr5_+_36611128 | 3.61 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr19_-_10243148 | 3.60 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr9_-_20372977 | 3.55 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr1_+_33969015 | 3.52 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr4_-_4780510 | 3.52 |
ENSDART00000109609
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr11_+_30513656 | 3.49 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr15_+_36156986 | 3.48 |
ENSDART00000059791
|
sst1.1
|
somatostatin 1, tandem duplicate 1 |
| chr19_+_10396042 | 3.46 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr20_+_27331008 | 3.45 |
ENSDART00000141486
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr23_-_44494401 | 3.42 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr10_+_29963518 | 3.42 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr17_-_17447899 | 3.39 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr19_+_17259912 | 3.36 |
ENSDART00000078951
|
MANEAL
|
si:ch211-30b16.2 |
| chr12_-_22540943 | 3.35 |
ENSDART00000172310
|
zbtb4
|
zinc finger and BTB domain containing 4 |
| chr5_-_16983336 | 3.35 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr4_-_4706893 | 3.31 |
ENSDART00000093005
|
CABZ01020835.1
|
|
| chr1_+_49814461 | 3.30 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr6_+_13920479 | 3.19 |
ENSDART00000155480
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr15_-_18574716 | 3.15 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr5_+_27583445 | 3.14 |
ENSDART00000136488
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr18_+_2837563 | 3.10 |
ENSDART00000171495
ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr2_+_31957554 | 3.10 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr11_+_30161699 | 3.09 |
ENSDART00000190504
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr18_+_49411417 | 3.05 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr17_-_20287530 | 3.01 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr4_-_27350820 | 3.01 |
ENSDART00000145806
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr11_-_24428237 | 3.00 |
ENSDART00000189107
ENSDART00000103752 |
dvl1b
|
dishevelled segment polarity protein 1b |
| chr18_-_23875219 | 3.00 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr20_-_7000225 | 3.00 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr24_-_11325849 | 2.96 |
ENSDART00000182485
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr4_-_12007404 | 2.95 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr15_+_15856178 | 2.94 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr13_-_30027730 | 2.92 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr2_+_47581997 | 2.91 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr3_-_62380146 | 2.90 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr1_+_14283692 | 2.87 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr14_+_790166 | 2.83 |
ENSDART00000123912
|
adra2da
|
adrenergic, alpha-2D-, receptor a |
| chr16_-_13004166 | 2.83 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr16_-_33059246 | 2.81 |
ENSDART00000171718
ENSDART00000168305 ENSDART00000166401 |
snap91
|
synaptosomal-associated protein 91 |
| chr4_-_211714 | 2.81 |
ENSDART00000172566
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr21_+_28958471 | 2.80 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr21_+_17110598 | 2.80 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr22_-_21150845 | 2.80 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr19_-_42651615 | 2.78 |
ENSDART00000123360
|
susd5
|
sushi domain containing 5 |
| chr23_-_8373676 | 2.77 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr2_+_45382433 | 2.77 |
ENSDART00000142251
|
wdr47a
|
WD repeat domain 47a |
| chr16_-_12953739 | 2.77 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr17_-_22067451 | 2.74 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr10_-_19801821 | 2.72 |
ENSDART00000148013
|
gfra2b
|
GDNF family receptor alpha 2b |
| chr8_-_11640240 | 2.70 |
ENSDART00000091752
|
fnbp1a
|
formin binding protein 1a |
| chr18_-_46208581 | 2.66 |
ENSDART00000141278
|
si:ch211-14c7.2
|
si:ch211-14c7.2 |
| chr2_+_42724404 | 2.66 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr21_-_41305748 | 2.64 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr16_-_13662514 | 2.62 |
ENSDART00000146348
|
shisa7a
|
shisa family member 7a |
| chr25_+_19954576 | 2.62 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr14_-_34276574 | 2.62 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr15_-_12319065 | 2.62 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr23_-_26077038 | 2.60 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr4_+_10888762 | 2.55 |
ENSDART00000136049
|
syt10
|
synaptotagmin X |
| chr15_+_38859648 | 2.52 |
ENSDART00000141086
|
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr24_-_18179535 | 2.49 |
ENSDART00000186112
|
cntnap2a
|
contactin associated protein like 2a |
| chr1_+_25783801 | 2.49 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr6_-_26559921 | 2.48 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr22_-_20105969 | 2.48 |
ENSDART00000088687
|
rxfp3.2b
|
relaxin/insulin-like family peptide receptor 3.2b |
| chr7_-_56793739 | 2.48 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr13_+_24022963 | 2.48 |
ENSDART00000028285
|
pgbd5
|
piggyBac transposable element derived 5 |
| chr1_+_36436936 | 2.46 |
ENSDART00000124112
|
pou4f2
|
POU class 4 homeobox 2 |
| chr2_-_32352946 | 2.45 |
ENSDART00000144870
ENSDART00000077151 |
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr13_+_12299997 | 2.45 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr8_-_23328264 | 2.45 |
ENSDART00000131934
|
nol4la
|
nucleolar protein 4-like a |
| chr17_+_38262408 | 2.45 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr14_-_7885707 | 2.43 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr21_+_23953181 | 2.43 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr1_+_8662530 | 2.43 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr10_+_22012389 | 2.42 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr7_+_10701770 | 2.40 |
ENSDART00000167323
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr12_-_48943467 | 2.39 |
ENSDART00000191829
|
CABZ01092907.1
|
|
| chr1_+_33383971 | 2.38 |
ENSDART00000150043
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr1_-_7917062 | 2.34 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr18_-_44610992 | 2.32 |
ENSDART00000125968
ENSDART00000185836 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_-_71460556 | 2.31 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr21_+_30194904 | 2.31 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr13_+_12120664 | 2.30 |
ENSDART00000130007
ENSDART00000079443 |
gabra2a
|
gamma-aminobutyric acid type A receptor alpha2 subunit a |
| chr3_+_46628885 | 2.30 |
ENSDART00000006602
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr19_+_15571290 | 2.29 |
ENSDART00000131134
|
foxo6b
|
forkhead box O6 b |
| chr8_+_49778756 | 2.29 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr9_-_23217196 | 2.28 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr10_+_41986685 | 2.28 |
ENSDART00000086165
ENSDART00000171983 |
setd1ba
|
SET domain containing 1B, a |
| chr15_+_33070939 | 2.27 |
ENSDART00000164928
|
mab21l1
|
mab-21-like 1 |
| chr21_+_6751405 | 2.27 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr8_+_10561922 | 2.22 |
ENSDART00000133348
|
fam19a5l
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5-like |
| chr11_-_36963988 | 2.21 |
ENSDART00000168288
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr13_-_20518632 | 2.19 |
ENSDART00000165310
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr18_-_23875370 | 2.19 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_-_22834824 | 2.19 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr19_-_9882821 | 2.18 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr17_+_28340138 | 2.18 |
ENSDART00000033943
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr14_+_8275115 | 2.18 |
ENSDART00000129055
|
nrg2b
|
neuregulin 2b |
| chr17_+_52822831 | 2.18 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr4_+_9279784 | 2.16 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr21_-_10446405 | 2.16 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr8_+_49778486 | 2.15 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr17_+_15535501 | 2.15 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr9_-_6661657 | 2.14 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr11_-_3959477 | 2.13 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr7_+_25858380 | 2.13 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr5_-_48285756 | 2.12 |
ENSDART00000183495
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr6_+_33537267 | 2.08 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr7_+_10701938 | 2.07 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_+_33383644 | 2.06 |
ENSDART00000187194
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr20_+_52774730 | 2.04 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr24_-_31846366 | 2.02 |
ENSDART00000155295
|
steap2
|
STEAP family member 2, metalloreductase |
| chr5_-_34185497 | 2.02 |
ENSDART00000146321
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr21_-_25522510 | 2.00 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr23_+_2560005 | 2.00 |
ENSDART00000186906
|
GGT7
|
gamma-glutamyltransferase 7 |
| chr21_+_6780340 | 1.98 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr5_-_28915130 | 1.98 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr11_+_19433936 | 1.98 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr2_+_21090317 | 1.98 |
ENSDART00000109568
ENSDART00000139633 |
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr5_-_34185115 | 1.96 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr5_+_3501859 | 1.94 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr7_-_38340674 | 1.91 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr11_+_30161168 | 1.91 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr8_-_17771755 | 1.90 |
ENSDART00000063592
|
prkcz
|
protein kinase C, zeta |
| chr8_+_1082100 | 1.90 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr1_+_6171585 | 1.89 |
ENSDART00000024358
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr13_+_12739283 | 1.89 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr11_+_34824099 | 1.88 |
ENSDART00000037017
ENSDART00000146944 |
slc38a3a
|
solute carrier family 38, member 3a |
| chr2_-_10188598 | 1.88 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr2_-_24603325 | 1.88 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr7_-_49800755 | 1.88 |
ENSDART00000180072
|
fjx1
|
four jointed box 1 |
| chr19_+_41006975 | 1.86 |
ENSDART00000138555
ENSDART00000049842 |
casd1
|
CAS1 domain containing 1 |
| chr12_-_14922955 | 1.85 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr20_-_47731768 | 1.84 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr6_+_4872883 | 1.83 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr1_-_8917902 | 1.83 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr16_+_1802307 | 1.82 |
ENSDART00000180026
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr9_-_43538328 | 1.81 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr3_+_14768364 | 1.81 |
ENSDART00000090235
ENSDART00000139001 |
nfixb
|
nuclear factor I/Xb |
| chr9_+_32978302 | 1.81 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr7_-_74090168 | 1.80 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr25_-_36282539 | 1.80 |
ENSDART00000073398
|
slc7a10b
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10b |
| chr18_-_46684352 | 1.79 |
ENSDART00000167520
|
pid1
|
phosphotyrosine interaction domain containing 1 |
| chr11_-_37589293 | 1.78 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr20_+_20637866 | 1.78 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr12_+_9703172 | 1.78 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr23_+_13124085 | 1.78 |
ENSDART00000139475
|
samd10b
|
sterile alpha motif domain containing 10b |
| chr4_-_17055782 | 1.77 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr24_-_11050070 | 1.77 |
ENSDART00000191320
ENSDART00000030409 ENSDART00000191248 ENSDART00000191943 |
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr5_-_43682930 | 1.77 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr8_-_5267442 | 1.77 |
ENSDART00000155816
|
ppp2r2aa
|
protein phosphatase 2, regulatory subunit B, alpha a |
| chr7_+_20629411 | 1.76 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr1_+_37195465 | 1.74 |
ENSDART00000043855
ENSDART00000192580 ENSDART00000181666 |
dclk2a
|
doublecortin-like kinase 2a |
| chr21_+_29227224 | 1.73 |
ENSDART00000013641
|
adra1ba
|
adrenoceptor alpha 1Ba |
| chr14_+_19258702 | 1.73 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr6_-_39489190 | 1.73 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr2_+_25929619 | 1.72 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr20_-_8419057 | 1.71 |
ENSDART00000145841
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr23_-_16692312 | 1.70 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr15_-_33734105 | 1.70 |
ENSDART00000172729
ENSDART00000172045 |
trmt9b
|
tRNA methyltransferase 9B |
| chr18_-_9046805 | 1.69 |
ENSDART00000134224
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr5_-_22952156 | 1.67 |
ENSDART00000111146
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr3_-_19091024 | 1.66 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr6_-_7208119 | 1.66 |
ENSDART00000105148
ENSDART00000185846 |
hs6st3a
|
heparan sulfate 6-O-sulfotransferase 3a |
| chr4_+_7888047 | 1.65 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr18_-_39787040 | 1.65 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr1_+_37195919 | 1.65 |
ENSDART00000159684
ENSDART00000172742 ENSDART00000158395 |
dclk2a
|
doublecortin-like kinase 2a |
| chr15_-_19128705 | 1.65 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr5_-_32092856 | 1.65 |
ENSDART00000086181
ENSDART00000181677 |
cabp7b
|
calcium binding protein 7b |
| chr8_+_14710542 | 1.65 |
ENSDART00000132899
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr19_+_33732487 | 1.64 |
ENSDART00000010294
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_+_33732188 | 1.63 |
ENSDART00000151192
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr18_+_16125852 | 1.63 |
ENSDART00000061106
|
bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr19_-_10395683 | 1.62 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr17_-_45552602 | 1.62 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr1_-_57280585 | 1.60 |
ENSDART00000152220
|
si:dkey-27j5.5
|
si:dkey-27j5.5 |
| chr1_-_45633955 | 1.60 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr1_-_22512063 | 1.60 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr15_-_12229874 | 1.59 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr4_+_2482046 | 1.56 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr21_-_27185915 | 1.56 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr25_+_1591964 | 1.53 |
ENSDART00000093277
|
ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr13_-_20519001 | 1.52 |
ENSDART00000168955
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr15_-_11681653 | 1.52 |
ENSDART00000180160
|
fkrp
|
fukutin related protein |
| chr9_+_25777270 | 1.52 |
ENSDART00000188547
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr11_+_34824262 | 1.52 |
ENSDART00000103157
|
slc38a3a
|
solute carrier family 38, member 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of arnt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 2.2 | 6.6 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 1.2 | 3.7 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 1.2 | 3.5 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 1.1 | 3.4 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 1.0 | 4.9 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.9 | 3.8 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.9 | 4.6 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.9 | 4.6 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.9 | 3.4 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.7 | 2.9 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.7 | 2.2 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.7 | 6.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.6 | 2.8 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.6 | 1.7 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.5 | 1.6 | GO:2000257 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.5 | 1.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.5 | 2.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.5 | 2.0 | GO:0015677 | copper ion import(GO:0015677) |
| 0.5 | 2.5 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.5 | 8.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.5 | 4.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.5 | 1.4 | GO:0097037 | heme export(GO:0097037) |
| 0.5 | 1.4 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.5 | 4.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.5 | 1.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 3.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.4 | 3.0 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.4 | 3.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.4 | 1.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.4 | 2.3 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.4 | 1.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.4 | 4.5 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.4 | 1.5 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.3 | 1.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 2.3 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.3 | 2.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 2.3 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.3 | 1.3 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.3 | 1.0 | GO:2000638 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.3 | 2.5 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.3 | 1.5 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.3 | 6.0 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 2.4 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.3 | 0.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.3 | 0.8 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 2.4 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 | 1.6 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.3 | 1.8 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.3 | 0.8 | GO:0045191 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.3 | 2.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.3 | 1.8 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 1.7 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 0.7 | GO:0042220 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.2 | 1.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 5.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 0.9 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.2 | 2.3 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.2 | 2.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 0.9 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 1.5 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 1.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 2.8 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.2 | 0.6 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.2 | 0.8 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.2 | 1.6 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 1.8 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 4.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 1.9 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.2 | 0.6 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.2 | 1.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 1.8 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 1.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 2.1 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.2 | 0.9 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.2 | 5.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 0.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 2.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 2.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 0.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 1.8 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.2 | 5.0 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.9 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.6 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 1.9 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 2.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.7 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 1.8 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.1 | 0.7 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 4.8 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 1.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 3.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 1.9 | GO:1902866 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 1.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 1.3 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 3.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.0 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.7 | GO:0010269 | response to selenium ion(GO:0010269) positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 4.0 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.1 | 1.0 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.5 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.1 | 0.5 | GO:0051701 | interaction with host(GO:0051701) |
| 0.1 | 0.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 2.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.8 | GO:0048675 | axon extension(GO:0048675) |
| 0.1 | 0.3 | GO:0043587 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.1 | 3.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 2.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 1.7 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 1.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 2.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.3 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 2.8 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 2.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.5 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 2.3 | GO:0070142 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.1 | 3.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 1.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 2.0 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 1.2 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.1 | 3.0 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 2.0 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 3.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.0 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 1.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.7 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 3.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 1.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 3.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 3.8 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 3.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.6 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 4.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.7 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 2.2 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 0.8 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 2.7 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 0.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.7 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.4 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.9 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 3.0 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.0 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.0 | 0.6 | GO:0035269 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 4.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 2.5 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 9.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 4.1 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.1 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 1.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.4 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 2.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.2 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.3 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.3 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 7.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 3.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 6.1 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.5 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) |
| 0.0 | 2.0 | GO:0051403 | stress-activated MAPK cascade(GO:0051403) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 2.0 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.7 | GO:0072089 | stem cell proliferation(GO:0072089) |
| 0.0 | 0.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 9.6 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.7 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.9 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.8 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.7 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 1.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.9 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.1 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 1.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 4.0 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 1.0 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.9 | 5.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 23.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 2.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 1.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 2.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 3.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 2.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.3 | 2.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 1.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 1.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 1.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.9 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 0.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 2.3 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.2 | 4.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 3.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 1.8 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 6.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 4.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 4.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 3.3 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 5.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.6 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 9.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 16.2 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 3.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 4.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 8.2 | GO:0099503 | exocytic vesicle(GO:0070382) secretory vesicle(GO:0099503) |
| 0.0 | 0.9 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 15.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 2.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 2.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 3.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 4.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.9 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 27.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.7 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.2 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.9 | 5.2 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 2.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.8 | 6.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.7 | 2.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.7 | 4.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.6 | 9.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.6 | 3.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.6 | 3.4 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.5 | 2.0 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 3.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 2.8 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.4 | 1.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.4 | 1.3 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.4 | 3.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 3.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 5.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 1.0 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.3 | 1.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 1.8 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 0.7 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.2 | 1.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 3.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 1.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 1.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 5.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 2.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.3 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.2 | 0.6 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.2 | 1.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 0.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 3.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 2.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 0.6 | GO:0005333 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.2 | 1.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 2.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 2.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 3.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 3.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 4.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 2.9 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 12.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 2.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 2.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 2.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 3.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.2 | GO:0005272 | sodium channel activity(GO:0005272) cAMP binding(GO:0030552) |
| 0.1 | 1.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 3.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 3.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 4.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 4.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 3.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.7 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 4.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 8.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 3.6 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 0.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.0 | GO:0010851 | G-protein alpha-subunit binding(GO:0001965) cyclase regulator activity(GO:0010851) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.2 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 1.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.4 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 2.0 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 2.3 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 3.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.2 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 2.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 1.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 2.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 4.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 5.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 2.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.9 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.1 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 1.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.8 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 4.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0002061 | nucleobase binding(GO:0002054) uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 4.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 11.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 4.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 4.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 4.2 | GO:0001653 | peptide receptor activity(GO:0001653) G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 7.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 1.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 4.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 8.8 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 2.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 5.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 5.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 1.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 2.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 1.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 4.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 1.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 1.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 7.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 3.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |