Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
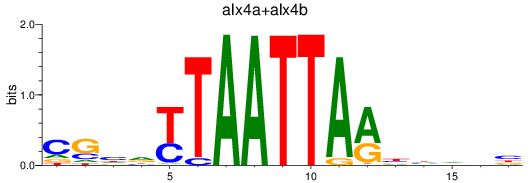
Results for alx4a+alx4b
Z-value: 0.65
Transcription factors associated with alx4a+alx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx4b
|
ENSDARG00000074442 | ALX homeobox 4b |
|
alx4a
|
ENSDARG00000088332 | ALX homeobox 4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx4b | dr11_v1_chr18_+_38321039_38321039 | -0.39 | 5.2e-01 | Click! |
| alx4a | dr11_v1_chr7_-_26924903_26924903 | 0.32 | 6.0e-01 | Click! |
Activity profile of alx4a+alx4b motif
Sorted Z-values of alx4a+alx4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_9669717 | 0.73 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr17_-_12336987 | 0.68 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr5_-_23280098 | 0.62 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr2_+_50608099 | 0.51 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr12_+_18578597 | 0.51 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr8_+_7359294 | 0.49 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr23_-_3681026 | 0.48 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr4_+_21129752 | 0.47 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr2_-_38000276 | 0.46 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr18_+_21408794 | 0.44 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr3_+_24361096 | 0.44 |
ENSDART00000132387
|
pvalb6
|
parvalbumin 6 |
| chr17_+_15433671 | 0.43 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr11_+_30057762 | 0.42 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr16_-_28856112 | 0.41 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr17_+_15433518 | 0.40 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr16_-_12173399 | 0.39 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr21_+_28958471 | 0.39 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr19_-_9867001 | 0.39 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr19_-_20113696 | 0.39 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr20_-_29864390 | 0.38 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr11_-_1509773 | 0.38 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr14_-_34276574 | 0.37 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr7_+_39634873 | 0.36 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr9_-_35334642 | 0.36 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr21_+_31150773 | 0.36 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr15_-_44601331 | 0.36 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr16_-_12173554 | 0.35 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr19_-_26869103 | 0.35 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr8_+_49778486 | 0.35 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr11_-_2131280 | 0.35 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr21_+_31150438 | 0.35 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr5_+_64739762 | 0.35 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr6_+_27151940 | 0.34 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr15_-_33925851 | 0.34 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr6_-_11768198 | 0.33 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr8_-_4596662 | 0.33 |
ENSDART00000138199
|
sept5a
|
septin 5a |
| chr8_+_49778756 | 0.32 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr6_-_40029423 | 0.32 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr17_-_200316 | 0.32 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr3_-_58543658 | 0.32 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr14_-_49063157 | 0.32 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr9_+_54039006 | 0.31 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr20_+_19512727 | 0.30 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr10_-_27049170 | 0.30 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr13_-_29420885 | 0.30 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr16_+_34531486 | 0.30 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr25_-_19420949 | 0.30 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr5_-_40190949 | 0.29 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr3_+_62000822 | 0.29 |
ENSDART00000106680
|
rai1
|
retinoic acid induced 1 |
| chr14_-_4556896 | 0.29 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr22_-_20011476 | 0.29 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr6_+_3004972 | 0.29 |
ENSDART00000186750
ENSDART00000183862 ENSDART00000191485 ENSDART00000171014 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr18_+_2837563 | 0.29 |
ENSDART00000171495
ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr24_-_7697274 | 0.29 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr25_+_126174 | 0.29 |
ENSDART00000168913
|
cntn1a
|
contactin 1a |
| chr13_-_11644806 | 0.28 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr9_-_43538328 | 0.28 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr23_+_16638639 | 0.27 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr24_+_41940299 | 0.27 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr10_-_2524297 | 0.26 |
ENSDART00000192475
|
CU856539.1
|
|
| chr6_+_3166941 | 0.26 |
ENSDART00000168912
|
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr8_+_6863462 | 0.26 |
ENSDART00000064163
|
nefmb
|
neurofilament, medium polypeptide b |
| chr3_-_6519691 | 0.25 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr9_+_2574122 | 0.25 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr19_-_15397946 | 0.25 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr3_-_21280373 | 0.25 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr2_+_53204750 | 0.25 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr3_-_32818607 | 0.25 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr15_-_9272328 | 0.25 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr6_-_39489190 | 0.25 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr21_-_20939488 | 0.24 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr12_+_31735159 | 0.24 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr6_-_46941245 | 0.24 |
ENSDART00000037875
|
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr8_+_36500061 | 0.24 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr25_-_12902242 | 0.24 |
ENSDART00000164733
|
sept15
|
septin 15 |
| chr17_-_36896560 | 0.23 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr10_-_2524917 | 0.23 |
ENSDART00000188642
|
CU856539.1
|
|
| chr17_-_3190070 | 0.23 |
ENSDART00000115027
|
tmem151bb
|
transmembrane protein 151Bb |
| chr25_-_13569234 | 0.23 |
ENSDART00000189645
|
fa2h
|
fatty acid 2-hydroxylase |
| chr19_+_5480327 | 0.23 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr14_+_22591624 | 0.23 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr22_+_17828267 | 0.23 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr7_-_72261721 | 0.23 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr11_+_41242644 | 0.23 |
ENSDART00000172008
|
pax7a
|
paired box 7a |
| chr7_+_23515966 | 0.22 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr4_-_1801519 | 0.22 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr9_-_1703761 | 0.22 |
ENSDART00000144822
ENSDART00000137210 ENSDART00000135273 |
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr20_+_3108597 | 0.22 |
ENSDART00000133435
|
CEP170B (1 of many)
|
si:ch73-212j7.1 |
| chr7_+_34620418 | 0.22 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr20_+_66857 | 0.22 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr21_-_13123176 | 0.22 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr3_-_23407720 | 0.21 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr25_+_5821910 | 0.21 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr16_+_2820340 | 0.21 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr23_+_44741500 | 0.21 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr15_+_46108548 | 0.21 |
ENSDART00000099023
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr22_-_13466246 | 0.20 |
ENSDART00000134035
|
cntnap5b
|
contactin associated protein-like 5b |
| chr9_+_29643036 | 0.20 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr2_+_55916911 | 0.20 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr21_+_27513859 | 0.20 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr5_+_59392183 | 0.20 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr5_+_37903790 | 0.19 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr21_+_21195487 | 0.19 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr16_-_41004731 | 0.19 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr6_+_35362225 | 0.19 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr6_-_46053300 | 0.19 |
ENSDART00000169722
|
ca16b
|
carbonic anhydrase XVI b |
| chr7_+_46368520 | 0.19 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr12_-_31794908 | 0.19 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr3_+_33341640 | 0.19 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr9_-_14504834 | 0.19 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr3_+_32443395 | 0.19 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr5_-_41494831 | 0.18 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr4_-_2219705 | 0.18 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr25_+_22719852 | 0.18 |
ENSDART00000088422
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr2_-_30668580 | 0.18 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr5_+_4054704 | 0.18 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr19_-_25149598 | 0.18 |
ENSDART00000162917
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr25_-_27722614 | 0.18 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr20_+_32552912 | 0.17 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr14_+_24284756 | 0.17 |
ENSDART00000079164
|
klhl3
|
kelch-like family member 3 |
| chr25_-_25058508 | 0.17 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr18_+_24919614 | 0.17 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr8_+_29742237 | 0.17 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr23_-_16692312 | 0.17 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr8_-_30979494 | 0.17 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr21_+_33503835 | 0.17 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr10_+_15255198 | 0.17 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr20_+_591505 | 0.17 |
ENSDART00000046438
|
kcnk2b
|
potassium channel, subfamily K, member 2b |
| chr24_+_5912898 | 0.17 |
ENSDART00000132686
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr4_+_9400012 | 0.17 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr16_-_32837806 | 0.16 |
ENSDART00000003997
|
MCHR2
|
si:dkey-165n16.5 |
| chr15_-_20939579 | 0.16 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr14_-_25078569 | 0.16 |
ENSDART00000172802
ENSDART00000173345 ENSDART00000135004 |
matr3l1.1
|
matrin 3-like 1.1 |
| chr12_+_47698356 | 0.16 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr17_-_21418340 | 0.16 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr15_-_22074315 | 0.16 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr4_+_3980247 | 0.16 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr5_-_66397688 | 0.16 |
ENSDART00000161483
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr13_+_29773153 | 0.16 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr25_+_31276842 | 0.16 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr24_+_5912635 | 0.16 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr15_-_30505607 | 0.16 |
ENSDART00000155212
|
msi2b
|
musashi RNA-binding protein 2b |
| chr13_-_18695289 | 0.16 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr4_-_5019113 | 0.15 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr23_-_333457 | 0.15 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr8_-_12264486 | 0.15 |
ENSDART00000091612
ENSDART00000135812 |
dab2ipa
|
DAB2 interacting protein a |
| chr10_+_25219728 | 0.15 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr25_+_29161609 | 0.15 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr15_-_19051152 | 0.15 |
ENSDART00000186453
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr3_+_5575313 | 0.15 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr1_+_6172786 | 0.15 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr5_+_21144269 | 0.15 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr22_-_12160283 | 0.15 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr8_+_17775247 | 0.14 |
ENSDART00000112356
|
si:ch211-150o23.3
|
si:ch211-150o23.3 |
| chr10_+_42423318 | 0.14 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr1_+_59293873 | 0.14 |
ENSDART00000168036
|
rdh8b
|
retinol dehydrogenase 8b |
| chr6_+_11249706 | 0.14 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr25_-_13490744 | 0.14 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr1_-_50438247 | 0.14 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr10_-_5581487 | 0.14 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr5_+_24156170 | 0.14 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr1_-_46981134 | 0.14 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr15_-_25527580 | 0.14 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr3_-_6767956 | 0.14 |
ENSDART00000192556
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr5_+_7279104 | 0.14 |
ENSDART00000190014
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr21_+_13387965 | 0.14 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr8_+_27807266 | 0.14 |
ENSDART00000170037
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr13_+_7292061 | 0.13 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr5_+_1109098 | 0.13 |
ENSDART00000166268
|
LO017790.1
|
|
| chr5_-_55933420 | 0.13 |
ENSDART00000050966
|
slc25a46
|
solute carrier family 25, member 46 |
| chr9_-_32177117 | 0.13 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr14_+_45406299 | 0.13 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr13_+_6342410 | 0.13 |
ENSDART00000065228
|
csmd1a
|
CUB and Sushi multiple domains 1a |
| chr22_-_16443199 | 0.13 |
ENSDART00000006290
ENSDART00000193335 |
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr7_-_71829649 | 0.13 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr5_-_12219572 | 0.13 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr3_+_13637383 | 0.13 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr17_+_41992054 | 0.13 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr1_-_15797663 | 0.13 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr14_+_29550323 | 0.13 |
ENSDART00000157943
|
TENM2
|
si:dkey-34l15.2 |
| chr14_+_8940326 | 0.12 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr8_+_36503797 | 0.12 |
ENSDART00000184785
|
slc7a4
|
solute carrier family 7, member 4 |
| chr16_-_28658341 | 0.12 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr19_+_5291935 | 0.12 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr20_-_28404362 | 0.12 |
ENSDART00000055932
ENSDART00000188161 |
pigh
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr6_+_11250033 | 0.12 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr25_-_6432463 | 0.12 |
ENSDART00000110389
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr16_+_13818500 | 0.12 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr2_+_2223837 | 0.12 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr4_+_16885854 | 0.12 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr12_-_41662327 | 0.12 |
ENSDART00000191602
ENSDART00000170423 |
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr24_-_5911973 | 0.12 |
ENSDART00000077933
ENSDART00000077922 |
pimr64
|
Pim proto-oncogene, serine/threonine kinase, related 64 |
| chr7_-_57509447 | 0.12 |
ENSDART00000051973
ENSDART00000147036 |
sirt3
|
sirtuin 3 |
| chr24_-_25691020 | 0.12 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr5_+_36752943 | 0.12 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr18_-_48983690 | 0.12 |
ENSDART00000182359
|
FO681288.3
|
|
| chr4_+_6572364 | 0.12 |
ENSDART00000122574
|
ppp1r3aa
|
protein phosphatase 1, regulatory subunit 3Aa |
| chr12_+_15148618 | 0.12 |
ENSDART00000125552
|
adprh
|
ADP-ribosylarginine hydrolase |
| chr11_+_44356707 | 0.11 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr10_-_14556978 | 0.11 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx4a+alx4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.3 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.4 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.4 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.1 | 0.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.2 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.3 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.0 | 0.6 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.0 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.3 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.0 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.6 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.2 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 0.4 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.0 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |