Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
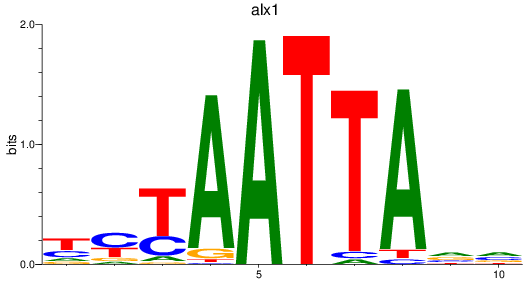
Results for alx1
Z-value: 1.09
Transcription factors associated with alx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx1
|
ENSDARG00000062824 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000110530 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000115230 | ALX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx1 | dr11_v1_chr18_+_16246806_16246806 | -0.12 | 8.5e-01 | Click! |
Activity profile of alx1 motif
Sorted Z-values of alx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 1.80 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr15_-_21877726 | 0.76 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr20_-_40755614 | 0.65 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr9_-_22821901 | 0.57 |
ENSDART00000101711
|
neb
|
nebulin |
| chr10_+_21867307 | 0.55 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr9_-_22834860 | 0.48 |
ENSDART00000146486
|
neb
|
nebulin |
| chr7_-_38792543 | 0.46 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr25_+_31267268 | 0.45 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr24_-_40744672 | 0.43 |
ENSDART00000160672
|
CU633479.1
|
|
| chr25_+_31277415 | 0.43 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr9_-_22822084 | 0.42 |
ENSDART00000142020
|
neb
|
nebulin |
| chr22_-_24818066 | 0.42 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr6_+_23887314 | 0.40 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr3_+_12816362 | 0.39 |
ENSDART00000163743
ENSDART00000170788 |
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr21_-_22737228 | 0.39 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr16_+_26777473 | 0.38 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr5_+_44805269 | 0.38 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr13_-_20381485 | 0.36 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr24_-_38110779 | 0.35 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr18_+_14564085 | 0.35 |
ENSDART00000009363
ENSDART00000141813 ENSDART00000136120 |
si:dkey-246g23.4
|
si:dkey-246g23.4 |
| chr19_-_42556086 | 0.35 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr2_-_1514001 | 0.33 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr21_-_23331619 | 0.33 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr10_+_6884123 | 0.32 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr6_-_53426773 | 0.32 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr5_-_23280098 | 0.32 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr23_-_27571667 | 0.31 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr9_-_49493305 | 0.31 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr25_+_3327071 | 0.31 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr13_-_50108337 | 0.30 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr20_-_23426339 | 0.30 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr15_-_2493771 | 0.30 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr10_+_6884627 | 0.30 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr18_+_15644559 | 0.29 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr14_+_22113331 | 0.29 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr6_+_55032439 | 0.29 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr5_+_37903790 | 0.28 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr19_+_15441022 | 0.28 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr15_-_2519640 | 0.27 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr13_+_2338761 | 0.27 |
ENSDART00000102791
ENSDART00000129911 |
klhl31
|
kelch-like family member 31 |
| chr2_-_30324610 | 0.27 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr2_-_15324837 | 0.26 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr5_+_28857969 | 0.26 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr18_+_8320165 | 0.26 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr13_+_38817871 | 0.26 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr8_+_45334255 | 0.25 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr7_+_20471315 | 0.25 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr10_+_43994471 | 0.24 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr12_-_14143344 | 0.24 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr2_-_30324297 | 0.24 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr22_-_23612854 | 0.24 |
ENSDART00000165885
|
cfhl5
|
complement factor H like 5 |
| chr5_+_51594209 | 0.24 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr16_-_17197546 | 0.23 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr10_-_7756865 | 0.23 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr22_+_16555939 | 0.23 |
ENSDART00000012604
|
pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr23_-_32157865 | 0.22 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_+_17695426 | 0.22 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr16_-_28658341 | 0.22 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr9_+_29548195 | 0.22 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr3_+_17537352 | 0.22 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr7_-_27685365 | 0.21 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr7_-_51773166 | 0.21 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr5_+_19314574 | 0.21 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr24_+_33392698 | 0.21 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr10_-_21362071 | 0.21 |
ENSDART00000125167
|
avd
|
avidin |
| chr24_+_17270129 | 0.20 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr2_-_37098785 | 0.20 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr16_-_16761164 | 0.20 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr15_-_20939579 | 0.20 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr2_-_51794472 | 0.20 |
ENSDART00000186652
|
BX908782.3
|
|
| chr5_-_56416188 | 0.20 |
ENSDART00000146773
|
acaca
|
acetyl-CoA carboxylase alpha |
| chr22_+_18477934 | 0.20 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr9_+_8396755 | 0.20 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr19_-_657439 | 0.19 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr22_-_10586191 | 0.19 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr10_-_21362320 | 0.19 |
ENSDART00000189789
|
avd
|
avidin |
| chr3_-_15080226 | 0.19 |
ENSDART00000109818
ENSDART00000139835 |
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr19_-_42571829 | 0.18 |
ENSDART00000102606
|
zgc:103438
|
zgc:103438 |
| chr23_+_40460333 | 0.18 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr20_-_7583486 | 0.18 |
ENSDART00000144729
|
usp24
|
ubiquitin specific peptidase 24 |
| chr7_-_34473389 | 0.18 |
ENSDART00000160532
|
si:zfos-1897c11.1
|
si:zfos-1897c11.1 |
| chr13_+_35637048 | 0.18 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr22_-_10459880 | 0.18 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr19_+_15440841 | 0.18 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr25_+_3306620 | 0.18 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr21_-_23110841 | 0.18 |
ENSDART00000147896
ENSDART00000003076 ENSDART00000184925 ENSDART00000190386 |
usp28
|
ubiquitin specific peptidase 28 |
| chr23_-_32162810 | 0.18 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr18_-_16801033 | 0.17 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr24_-_3783497 | 0.17 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr2_+_6253246 | 0.17 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr23_+_41679586 | 0.17 |
ENSDART00000067662
|
CU914487.1
|
|
| chr1_+_24387659 | 0.17 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr19_-_20403845 | 0.17 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr5_+_64732036 | 0.17 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr13_-_10945288 | 0.16 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr14_+_36521005 | 0.16 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr7_-_31759602 | 0.16 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr9_-_19161982 | 0.16 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr22_-_20924564 | 0.16 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr16_-_29387215 | 0.16 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr17_+_52612866 | 0.16 |
ENSDART00000182828
ENSDART00000191156 ENSDART00000188814 ENSDART00000109891 |
angel1
|
angel homolog 1 (Drosophila) |
| chr19_+_7152966 | 0.16 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr15_-_16098531 | 0.15 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr8_+_25034544 | 0.15 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr1_-_12109216 | 0.15 |
ENSDART00000079930
|
mttp
|
microsomal triglyceride transfer protein |
| chr13_+_38990939 | 0.15 |
ENSDART00000145979
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr10_-_25217347 | 0.15 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr9_-_31278048 | 0.15 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr5_-_56412262 | 0.15 |
ENSDART00000083079
|
acaca
|
acetyl-CoA carboxylase alpha |
| chr6_-_46861676 | 0.15 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr5_-_9216758 | 0.15 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr24_+_12835935 | 0.14 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr6_+_6828167 | 0.14 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr20_+_38032143 | 0.14 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr23_+_24272421 | 0.14 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr10_-_26744131 | 0.14 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr17_-_23416897 | 0.14 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr1_-_9249943 | 0.14 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr17_+_46818521 | 0.14 |
ENSDART00000156022
|
pimr14
|
Pim proto-oncogene, serine/threonine kinase, related 14 |
| chr1_-_55068941 | 0.14 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr1_-_513762 | 0.14 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr16_-_42056137 | 0.14 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr6_+_41191482 | 0.14 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr6_+_8626427 | 0.14 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr25_-_25384045 | 0.14 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr7_-_31759394 | 0.14 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr9_+_21146862 | 0.14 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr17_+_21295132 | 0.14 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr16_+_40563533 | 0.13 |
ENSDART00000190368
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr20_+_32118559 | 0.13 |
ENSDART00000026273
|
cd164
|
CD164 molecule, sialomucin |
| chr24_+_7336411 | 0.13 |
ENSDART00000191170
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr24_-_21914276 | 0.13 |
ENSDART00000128687
|
c1qtnf9
|
C1q and TNF related 9 |
| chr11_-_41853874 | 0.13 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr15_-_34458495 | 0.13 |
ENSDART00000059954
|
meox2a
|
mesenchyme homeobox 2a |
| chr24_-_29963858 | 0.13 |
ENSDART00000183442
|
CR352310.1
|
|
| chr1_-_44812014 | 0.13 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr24_+_21540842 | 0.13 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr8_-_28449782 | 0.13 |
ENSDART00000062702
|
cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr16_+_39159752 | 0.13 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr14_-_12307522 | 0.13 |
ENSDART00000163900
|
myot
|
myotilin |
| chr14_+_44794936 | 0.13 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr15_+_34988148 | 0.13 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr15_-_4969525 | 0.13 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr8_+_635704 | 0.12 |
ENSDART00000130358
|
csgalnact1b
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1b |
| chr12_-_30359498 | 0.12 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr16_+_27349585 | 0.12 |
ENSDART00000142573
|
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr9_-_50001606 | 0.12 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr9_+_50001746 | 0.12 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr8_+_3820134 | 0.12 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr14_-_2933185 | 0.12 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr20_+_46925581 | 0.12 |
ENSDART00000192531
|
si:ch73-21k16.5
|
si:ch73-21k16.5 |
| chr21_-_35419486 | 0.12 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr24_+_26895748 | 0.12 |
ENSDART00000089351
|
nceh1b.1
|
neutral cholesterol ester hydrolase 1b, tandem duplicate 1 |
| chr6_+_9204099 | 0.12 |
ENSDART00000150167
ENSDART00000115394 |
tbx19
|
T-box 19 |
| chr16_-_46579936 | 0.12 |
ENSDART00000166143
ENSDART00000127212 |
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr23_-_17003533 | 0.12 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr7_+_73397283 | 0.12 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr8_+_41037541 | 0.12 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr15_+_31344472 | 0.12 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr18_+_38807239 | 0.12 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr12_-_30359031 | 0.11 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr17_-_29119362 | 0.11 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr4_-_14315855 | 0.11 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr22_+_1170294 | 0.11 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr2_+_25198648 | 0.11 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr5_+_35786141 | 0.11 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr9_+_23224761 | 0.11 |
ENSDART00000142008
|
map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr2_+_37227011 | 0.11 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr8_-_45838277 | 0.11 |
ENSDART00000046064
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr19_+_46158078 | 0.11 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr22_-_8725768 | 0.11 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr8_-_46525092 | 0.11 |
ENSDART00000030482
|
sult1st2
|
sulfotransferase family 1, cytosolic sulfotransferase 2 |
| chr15_-_22074315 | 0.11 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr11_-_1550709 | 0.11 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr4_+_14900042 | 0.11 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr14_+_34492288 | 0.11 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr23_-_30954738 | 0.11 |
ENSDART00000188996
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr20_-_7582936 | 0.11 |
ENSDART00000083890
|
usp24
|
ubiquitin specific peptidase 24 |
| chr1_-_55248496 | 0.11 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr4_-_20292821 | 0.11 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr21_+_43404945 | 0.11 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr9_+_29548630 | 0.11 |
ENSDART00000132295
|
rnf17
|
ring finger protein 17 |
| chr24_+_5208171 | 0.11 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr4_+_12612723 | 0.11 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr20_+_4060839 | 0.11 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr12_+_48803098 | 0.10 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr6_+_16736871 | 0.10 |
ENSDART00000155471
|
pimr12
|
Pim proto-oncogene, serine/threonine kinase, related 12 |
| chr22_-_10486477 | 0.10 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr6_+_9870192 | 0.10 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr17_+_23298928 | 0.10 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr9_+_34641237 | 0.10 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr5_-_39698224 | 0.10 |
ENSDART00000076929
|
prkg2
|
protein kinase, cGMP-dependent, type II |
| chr24_-_25166720 | 0.10 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr24_+_2495197 | 0.10 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr20_-_16171297 | 0.10 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr2_+_58221163 | 0.10 |
ENSDART00000157939
|
FO704813.1
|
|
| chr5_+_57924611 | 0.10 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr8_+_20825987 | 0.10 |
ENSDART00000133309
|
si:ch211-133l5.4
|
si:ch211-133l5.4 |
| chr24_-_31452875 | 0.10 |
ENSDART00000187381
ENSDART00000185128 |
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr1_-_29139141 | 0.10 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 0.8 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.4 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.5 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.2 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 2.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 0.4 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.2 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.6 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.2 | GO:0065005 | plasma lipoprotein particle assembly(GO:0034377) protein-lipid complex assembly(GO:0065005) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.3 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 0.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.0 | 0.1 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.4 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.0 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.8 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.0 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.4 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.0 | 0.2 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.0 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |