Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
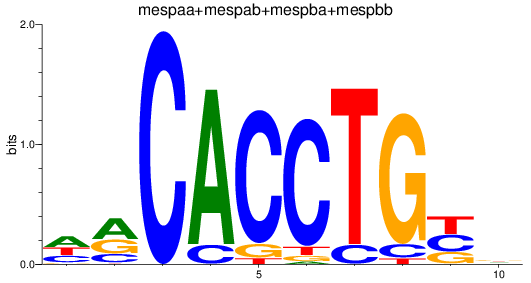
Results for TCF4_id4_mespaa+mespab+mespba+mespbb
Z-value: 1.48
Transcription factors associated with TCF4_id4_mespaa+mespab+mespba+mespbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
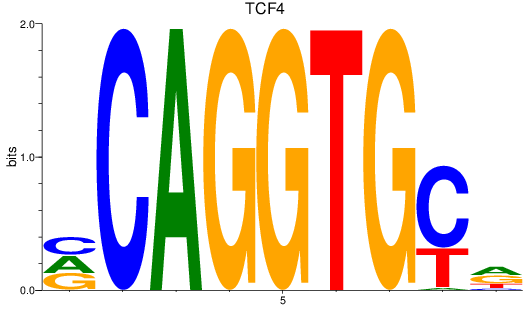
TCF4
|
ENSDARG00000107408 | transcription factor 4 |
|
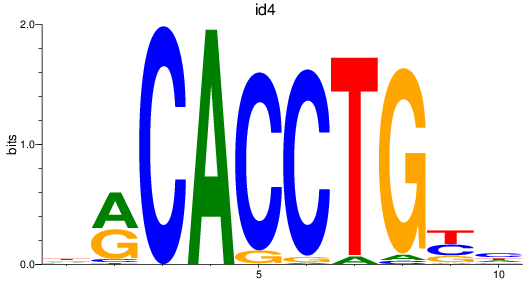
id4
|
ENSDARG00000045131 | inhibitor of DNA binding 4 |
|
mespaa
|
ENSDARG00000017078 | mesoderm posterior aa |
|
mespba
|
ENSDARG00000030347 | mesoderm posterior ba |
|
mespab
|
ENSDARG00000068761 | mesoderm posterior ab |
|
mespbb
|
ENSDARG00000097947 | mesoderm posterior bb |
|
mespba
|
ENSDARG00000110553 | mesoderm posterior ba |
|
mespaa
|
ENSDARG00000114890 | mesoderm posterior aa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mespaa | dr11_v1_chr7_+_15324830_15324830 | -0.82 | 9.0e-02 | Click! |
| id4 | dr11_v1_chr16_-_9980402_9980402 | -0.70 | 1.9e-01 | Click! |
| TCF4 | dr11_v1_chr21_+_1381276_1381276 | -0.44 | 4.5e-01 | Click! |
| mespbb | dr11_v1_chr25_-_11026907_11026907 | -0.42 | 4.8e-01 | Click! |
Activity profile of TCF4_id4_mespaa+mespab+mespba+mespbb motif
Sorted Z-values of TCF4_id4_mespaa+mespab+mespba+mespbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48312109 | 1.26 |
ENSDART00000161103
|
si:ch73-359m17.9
|
si:ch73-359m17.9 |
| chr16_-_36834505 | 1.07 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr20_-_42702832 | 1.01 |
ENSDART00000134689
ENSDART00000045816 |
plg
|
plasminogen |
| chr2_-_22688651 | 0.77 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr7_+_7048245 | 0.77 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr24_-_25691020 | 0.69 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr1_+_30422143 | 0.66 |
ENSDART00000033024
|
tmem41ab
|
transmembrane protein 41ab |
| chr7_+_39446247 | 0.64 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr9_+_310331 | 0.64 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr25_-_4146947 | 0.61 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr22_-_25612680 | 0.61 |
ENSDART00000114167
|
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr1_-_18811517 | 0.60 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr7_+_58751504 | 0.59 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr21_-_43666420 | 0.57 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr7_+_61184551 | 0.56 |
ENSDART00000190788
|
zgc:194930
|
zgc:194930 |
| chr11_-_37995501 | 0.56 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr3_+_42923275 | 0.55 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr21_-_30293224 | 0.55 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr21_-_34261677 | 0.55 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr25_-_32311048 | 0.54 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr10_-_25217347 | 0.54 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr6_+_29305190 | 0.54 |
ENSDART00000078647
|
si:ch211-201h21.5
|
si:ch211-201h21.5 |
| chr9_-_34269066 | 0.52 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr9_+_307863 | 0.52 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr5_-_38451082 | 0.51 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr15_-_43164591 | 0.51 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr16_+_52512025 | 0.50 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr13_-_21672131 | 0.50 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr2_-_50225411 | 0.49 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr6_-_40058686 | 0.49 |
ENSDART00000103240
|
uroc1
|
urocanate hydratase 1 |
| chr23_+_39854566 | 0.49 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr14_+_15155684 | 0.48 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr22_-_17677947 | 0.47 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr2_-_47620806 | 0.47 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr3_+_32526799 | 0.47 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr3_+_32526263 | 0.47 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr12_-_23365737 | 0.47 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr21_-_25756119 | 0.46 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr6_+_153146 | 0.46 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr1_+_24387659 | 0.46 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr4_-_77557279 | 0.44 |
ENSDART00000180113
|
AL935186.10
|
|
| chr20_+_26556372 | 0.44 |
ENSDART00000187179
ENSDART00000157291 |
irf4b
|
interferon regulatory factor 4b |
| chr2_+_6253246 | 0.44 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr7_+_6969909 | 0.43 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr14_+_11458044 | 0.43 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr23_+_553396 | 0.43 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr15_-_3277635 | 0.43 |
ENSDART00000189094
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr3_+_14641962 | 0.43 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr2_+_11031360 | 0.43 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr9_+_7358749 | 0.42 |
ENSDART00000081660
|
ihha
|
Indian hedgehog homolog a |
| chr6_+_60112200 | 0.42 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr9_-_39005317 | 0.42 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr17_-_30473374 | 0.41 |
ENSDART00000155021
|
si:ch211-175f11.5
|
si:ch211-175f11.5 |
| chr12_+_13091842 | 0.40 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr17_-_2596125 | 0.40 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr21_-_26114886 | 0.40 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr9_-_22892838 | 0.39 |
ENSDART00000143888
|
neb
|
nebulin |
| chr2_-_41942666 | 0.39 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr16_-_42066523 | 0.38 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr23_+_6077503 | 0.38 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr22_+_661711 | 0.38 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr2_+_27855346 | 0.38 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr14_+_22297257 | 0.37 |
ENSDART00000113676
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr5_-_69212184 | 0.37 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr22_+_25249193 | 0.37 |
ENSDART00000171851
|
si:ch211-226h8.11
|
si:ch211-226h8.11 |
| chr19_+_48117995 | 0.37 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr4_+_72797711 | 0.37 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr6_+_45918981 | 0.36 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr17_+_53294228 | 0.36 |
ENSDART00000158172
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr22_+_661505 | 0.36 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr20_-_21672970 | 0.36 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr10_+_38775959 | 0.36 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr10_+_38775408 | 0.36 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr22_+_25248961 | 0.36 |
ENSDART00000143079
|
si:ch211-226h8.11
|
si:ch211-226h8.11 |
| chr22_-_14128716 | 0.36 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr10_+_34001444 | 0.35 |
ENSDART00000149934
|
kl
|
klotho |
| chr2_-_35566938 | 0.35 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr14_-_46198373 | 0.35 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr12_+_46543572 | 0.35 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr7_+_57089354 | 0.35 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr10_-_13116337 | 0.34 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr24_+_35564668 | 0.34 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr2_+_57801960 | 0.33 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr7_+_20471315 | 0.33 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr14_+_11457500 | 0.33 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr2_+_17181777 | 0.32 |
ENSDART00000112063
|
ptger4c
|
prostaglandin E receptor 4 (subtype EP4) c |
| chr8_+_15254564 | 0.32 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr11_+_329687 | 0.32 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr15_-_3282220 | 0.32 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr12_+_6041575 | 0.32 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr15_+_29025090 | 0.32 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr2_+_27855102 | 0.31 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr7_+_61184104 | 0.31 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr2_+_394166 | 0.31 |
ENSDART00000155733
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr5_-_69948099 | 0.31 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr20_-_27325258 | 0.31 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr3_+_25849560 | 0.31 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr18_+_45666489 | 0.30 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr23_-_19684721 | 0.30 |
ENSDART00000184325
|
zgc:193598
|
zgc:193598 |
| chr13_-_12581388 | 0.30 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr15_-_33964897 | 0.30 |
ENSDART00000172075
ENSDART00000158126 ENSDART00000160456 |
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr20_+_26556174 | 0.30 |
ENSDART00000138492
|
irf4b
|
interferon regulatory factor 4b |
| chr2_-_24402341 | 0.30 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr13_-_34683370 | 0.30 |
ENSDART00000113661
|
kif16bb
|
kinesin family member 16Bb |
| chr2_+_11028923 | 0.30 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr10_-_15053507 | 0.30 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr10_+_3520256 | 0.29 |
ENSDART00000003242
|
zgc:123275
|
zgc:123275 |
| chr5_-_37900350 | 0.29 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr6_+_6780873 | 0.29 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr17_-_2595736 | 0.29 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr15_+_29024895 | 0.29 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr19_+_19767567 | 0.28 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr7_-_50367326 | 0.28 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr7_+_41303572 | 0.28 |
ENSDART00000012692
|
rgs9bp
|
regulator of G protein signaling 9 binding protein |
| chr3_+_17456428 | 0.28 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr1_-_12027 | 0.28 |
ENSDART00000164359
|
rpl24
|
ribosomal protein L24 |
| chr4_-_68563862 | 0.28 |
ENSDART00000182970
|
BX548011.2
|
|
| chr14_-_33277743 | 0.27 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr3_+_22273123 | 0.27 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr21_-_25741096 | 0.27 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr6_-_39270851 | 0.27 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr11_+_11175814 | 0.27 |
ENSDART00000183864
|
CR847973.1
|
|
| chr24_-_25244637 | 0.27 |
ENSDART00000153798
|
hhla2b.2
|
HERV-H LTR-associating 2b, tandem duplicate 2 |
| chr17_-_25331439 | 0.27 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr19_-_10771558 | 0.27 |
ENSDART00000085165
|
slc9a3.2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 2 |
| chr6_+_42338309 | 0.26 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr16_-_12723738 | 0.26 |
ENSDART00000080414
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr7_+_30392613 | 0.26 |
ENSDART00000075508
|
lipca
|
lipase, hepatic a |
| chr8_+_45334255 | 0.26 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr7_-_53117131 | 0.26 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr5_-_37047147 | 0.26 |
ENSDART00000097712
ENSDART00000158974 |
ftr99
|
finTRIM family, member 99 |
| chr7_-_55648336 | 0.26 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr21_-_7928101 | 0.26 |
ENSDART00000151543
ENSDART00000114982 |
f2rl1.2
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 2 |
| chr21_-_25741411 | 0.26 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr1_+_12009673 | 0.25 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr23_-_32156278 | 0.25 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr2_+_11029138 | 0.25 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr7_+_29951997 | 0.25 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr7_-_48251234 | 0.25 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr24_-_33703504 | 0.25 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr12_+_45676667 | 0.25 |
ENSDART00000016553
|
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr19_+_15441022 | 0.24 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr21_-_40630650 | 0.24 |
ENSDART00000172706
|
pcyt1bb
|
phosphate cytidylyltransferase 1, choline, beta b |
| chr3_+_36424055 | 0.24 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr16_-_42056137 | 0.24 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr23_-_29824146 | 0.24 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr3_+_62161184 | 0.24 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr20_+_10538025 | 0.24 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr7_-_50367642 | 0.24 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr6_-_609880 | 0.24 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr15_+_43906043 | 0.24 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr5_-_1962500 | 0.24 |
ENSDART00000150163
|
rplp0
|
ribosomal protein, large, P0 |
| chr25_-_17395315 | 0.24 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr10_-_26744131 | 0.24 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr11_-_25853212 | 0.24 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr24_+_10898671 | 0.23 |
ENSDART00000106272
|
si:dkey-37o8.1
|
si:dkey-37o8.1 |
| chr5_-_6567464 | 0.23 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr20_-_53624645 | 0.23 |
ENSDART00000083427
ENSDART00000152920 |
slc25a29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr4_+_14981854 | 0.23 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr19_+_25504645 | 0.23 |
ENSDART00000143292
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr16_+_46725087 | 0.23 |
ENSDART00000008920
|
rab11al
|
RAB11a, member RAS oncogene family, like |
| chr21_-_43636595 | 0.23 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr16_-_47381519 | 0.23 |
ENSDART00000032188
ENSDART00000150136 |
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr1_-_29747702 | 0.23 |
ENSDART00000133225
ENSDART00000189670 |
spp2
|
secreted phosphoprotein 2 |
| chr14_+_26546175 | 0.23 |
ENSDART00000105932
|
si:dkeyp-110e4.11
|
si:dkeyp-110e4.11 |
| chr7_+_14005111 | 0.23 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr14_-_83154 | 0.23 |
ENSDART00000187097
|
CABZ01086812.1
|
|
| chr24_-_6038025 | 0.22 |
ENSDART00000077819
ENSDART00000139216 |
ftr61
|
finTRIM family, member 61 |
| chr5_-_23715861 | 0.22 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr3_-_61205711 | 0.22 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr1_-_20593778 | 0.22 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr22_-_17256573 | 0.22 |
ENSDART00000136119
ENSDART00000062891 |
nphs2
|
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr7_+_24628546 | 0.22 |
ENSDART00000173745
|
MSMP
|
si:ch211-69e13.6 |
| chr25_-_17587785 | 0.22 |
ENSDART00000073679
ENSDART00000146851 |
zgc:66449
|
zgc:66449 |
| chr16_-_17713859 | 0.22 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr4_-_948776 | 0.22 |
ENSDART00000023483
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr10_+_31809226 | 0.22 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr7_+_69841017 | 0.22 |
ENSDART00000169107
|
FO818704.1
|
|
| chr1_-_34450622 | 0.21 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr17_+_53292215 | 0.21 |
ENSDART00000170686
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr11_-_34097418 | 0.21 |
ENSDART00000028301
ENSDART00000138579 |
tdo2b
|
tryptophan 2,3-dioxygenase b |
| chr7_+_57088920 | 0.21 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr7_+_67699009 | 0.21 |
ENSDART00000192810
|
zgc:162592
|
zgc:162592 |
| chr6_-_13498745 | 0.21 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr21_-_20725853 | 0.21 |
ENSDART00000114502
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr22_+_22021936 | 0.21 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr5_-_64355227 | 0.21 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr19_+_2590182 | 0.21 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr3_+_14388010 | 0.21 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr8_+_32719930 | 0.21 |
ENSDART00000145362
|
hmcn2
|
hemicentin 2 |
| chr6_+_42918933 | 0.21 |
ENSDART00000064896
|
gnat1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| chr15_+_20239141 | 0.21 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr7_+_27455321 | 0.21 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr17_-_36860988 | 0.21 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr7_+_55112922 | 0.21 |
ENSDART00000073549
|
snai3
|
snail family zinc finger 3 |
| chr5_-_23855447 | 0.20 |
ENSDART00000051541
|
gbgt1l3
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 3 |
| chr8_+_53051701 | 0.20 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr10_-_35149513 | 0.20 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr8_-_25690966 | 0.20 |
ENSDART00000033701
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr13_-_9442942 | 0.20 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF4_id4_mespaa+mespab+mespba+mespbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 0.8 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.2 | 1.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 0.5 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 0.9 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.4 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 2.2 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.9 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.7 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.3 | GO:0030237 | female sex determination(GO:0030237) |
| 0.1 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.1 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.5 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.2 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.2 | GO:0090008 | convergent extension involved in nephron morphogenesis(GO:0072045) hypoblast development(GO:0090008) |
| 0.1 | 0.6 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.2 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.5 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.3 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.1 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.1 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.4 | GO:0042436 | tryptophan metabolic process(GO:0006568) tryptophan catabolic process(GO:0006569) indolalkylamine metabolic process(GO:0006586) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.2 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.2 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0002902 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.1 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:2000648 | neuroblast division(GO:0055057) positive regulation of stem cell proliferation(GO:2000648) |
| 0.0 | 0.3 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.6 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.8 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.0 | 0.1 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.1 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.4 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0032262 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 0.1 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.1 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.1 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.2 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.5 | GO:0009994 | oocyte differentiation(GO:0009994) |
| 0.0 | 0.6 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.2 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.0 | 0.2 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.1 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.0 | 0.2 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.0 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.2 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.1 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:0089718 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.0 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 1.0 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.0 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.1 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.2 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.0 | 0.0 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.0 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.7 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 1.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.9 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.5 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.2 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.2 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.2 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.0 | 0.3 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.3 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.6 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.0 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |