Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
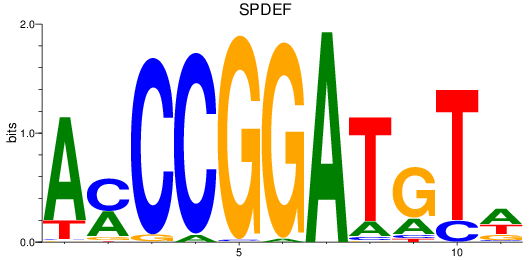
Results for SPDEF
Z-value: 1.23
Transcription factors associated with SPDEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPDEF
|
ENSDARG00000029930 | SAM pointed domain containing ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPDEF | dr11_v1_chr6_-_54290227_54290227 | 0.51 | 3.8e-01 | Click! |
Activity profile of SPDEF motif
Sorted Z-values of SPDEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_37118547 | 0.25 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr17_-_27382826 | 0.25 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr17_+_33375469 | 0.23 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr8_+_23726244 | 0.22 |
ENSDART00000132734
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr5_+_44346691 | 0.22 |
ENSDART00000034523
|
tars
|
threonyl-tRNA synthetase |
| chr8_+_23726708 | 0.21 |
ENSDART00000142395
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr20_+_52492192 | 0.20 |
ENSDART00000057986
|
TSTA3 (1 of many)
|
zgc:100864 |
| chr8_-_17184482 | 0.19 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr21_-_19828423 | 0.19 |
ENSDART00000037664
|
nnt
|
nicotinamide nucleotide transhydrogenase |
| chr20_+_39685572 | 0.19 |
ENSDART00000050729
|
hddc2
|
HD domain containing 2 |
| chr22_+_5663529 | 0.19 |
ENSDART00000106141
|
tma7
|
translation machinery associated 7 homolog |
| chr2_+_4383061 | 0.19 |
ENSDART00000163986
|
wacb
|
WW domain containing adaptor with coiled-coil b |
| chr4_-_18954001 | 0.18 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr7_+_59649399 | 0.18 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr15_+_46313082 | 0.18 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr22_-_5663354 | 0.18 |
ENSDART00000081774
|
ccdc51
|
coiled-coil domain containing 51 |
| chr23_-_33775145 | 0.18 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr21_-_30994577 | 0.18 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr13_-_12494575 | 0.18 |
ENSDART00000137761
|
si:dkey-20i10.7
|
si:dkey-20i10.7 |
| chr23_+_16928506 | 0.18 |
ENSDART00000162292
ENSDART00000080625 |
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr3_+_30980852 | 0.17 |
ENSDART00000028529
|
prf1.1
|
perforin 1.1 |
| chr19_+_9455218 | 0.17 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr19_+_34230108 | 0.17 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr23_-_40250886 | 0.17 |
ENSDART00000041912
|
tgm1l3
|
transglutaminase 1 like 3 |
| chr6_-_29288155 | 0.17 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr2_+_49457626 | 0.17 |
ENSDART00000129967
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr3_-_40276057 | 0.16 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr21_-_40938382 | 0.16 |
ENSDART00000008593
|
yipf5
|
Yip1 domain family, member 5 |
| chr11_-_15296805 | 0.16 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr23_+_9867483 | 0.16 |
ENSDART00000023099
|
slc16a7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2) |
| chr24_-_32522587 | 0.16 |
ENSDART00000048968
ENSDART00000143781 |
EIF1B
|
zgc:56676 |
| chr2_+_49457449 | 0.16 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr6_+_4387150 | 0.15 |
ENSDART00000181283
|
rbm26
|
RNA binding motif protein 26 |
| chr4_+_9612574 | 0.15 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr19_-_35428815 | 0.15 |
ENSDART00000169006
ENSDART00000003167 |
ak2
|
adenylate kinase 2 |
| chr8_-_41279326 | 0.15 |
ENSDART00000075491
|
pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr21_-_36619599 | 0.15 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr4_-_22749553 | 0.15 |
ENSDART00000040033
|
nup107
|
nucleoporin 107 |
| chr24_-_10897511 | 0.15 |
ENSDART00000145593
ENSDART00000102484 ENSDART00000066784 |
fam49bb
|
family with sequence similarity 49, member Bb |
| chr12_+_46543572 | 0.15 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr21_-_32436679 | 0.15 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr20_+_39344889 | 0.14 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr5_+_26795773 | 0.14 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr22_-_20950448 | 0.14 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr19_-_34089205 | 0.14 |
ENSDART00000163618
ENSDART00000161666 |
rp9
|
RP9, pre-mRNA splicing factor |
| chr16_+_12812472 | 0.14 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr2_-_20052561 | 0.14 |
ENSDART00000100133
|
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr20_-_20611063 | 0.14 |
ENSDART00000063492
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr16_+_54829574 | 0.14 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr23_-_24234371 | 0.14 |
ENSDART00000124539
|
ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr13_-_5569562 | 0.14 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr1_+_49435017 | 0.14 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr13_+_31545812 | 0.13 |
ENSDART00000076527
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr11_-_15297129 | 0.13 |
ENSDART00000167985
ENSDART00000163031 ENSDART00000165010 ENSDART00000171250 ENSDART00000010684 ENSDART00000191714 ENSDART00000191164 |
rpn2
|
ribophorin II |
| chr9_-_28255029 | 0.13 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr19_+_43004408 | 0.13 |
ENSDART00000038230
|
snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr23_+_4260458 | 0.13 |
ENSDART00000103747
|
srsf6a
|
serine/arginine-rich splicing factor 6a |
| chr5_-_23117078 | 0.13 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr19_+_7735157 | 0.13 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr8_-_53680653 | 0.13 |
ENSDART00000164739
|
wnt5a
|
wingless-type MMTV integration site family, member 5a |
| chr1_+_47165842 | 0.13 |
ENSDART00000053152
ENSDART00000167051 |
cbr1
|
carbonyl reductase 1 |
| chr12_+_29240124 | 0.13 |
ENSDART00000053761
ENSDART00000130172 |
bms1
|
BMS1 ribosome biogenesis factor |
| chr12_-_14211293 | 0.13 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr7_-_57637779 | 0.13 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr23_-_30764319 | 0.13 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr7_+_59649746 | 0.13 |
ENSDART00000181067
|
rpl34
|
ribosomal protein L34 |
| chr14_-_5407555 | 0.13 |
ENSDART00000001424
|
pcgf1
|
polycomb group ring finger 1 |
| chr18_-_22735002 | 0.13 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr13_+_31545530 | 0.13 |
ENSDART00000164590
ENSDART00000178460 ENSDART00000185503 |
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr11_-_16975190 | 0.13 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr2_+_42212948 | 0.13 |
ENSDART00000147019
|
si:ch211-235o23.1
|
si:ch211-235o23.1 |
| chr2_-_2642476 | 0.13 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr18_+_29898955 | 0.13 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr3_-_3398383 | 0.13 |
ENSDART00000047865
|
si:dkey-46g23.2
|
si:dkey-46g23.2 |
| chr3_-_27065477 | 0.13 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr16_-_44673851 | 0.12 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr20_-_23946296 | 0.12 |
ENSDART00000143005
|
mdn1
|
midasin AAA ATPase 1 |
| chr1_+_46598502 | 0.12 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr6_-_18531349 | 0.12 |
ENSDART00000160693
ENSDART00000169780 |
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr25_-_17239008 | 0.12 |
ENSDART00000151965
ENSDART00000152106 ENSDART00000152107 |
ccnd2a
|
cyclin D2, a |
| chr16_+_12812214 | 0.12 |
ENSDART00000124875
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr2_-_45510699 | 0.12 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr14_-_10617923 | 0.12 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr21_+_26539157 | 0.12 |
ENSDART00000021121
|
stx5al
|
syntaxin 5A, like |
| chr24_-_26369185 | 0.12 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr18_+_6638726 | 0.11 |
ENSDART00000142755
ENSDART00000167781 |
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr2_-_43130812 | 0.11 |
ENSDART00000039079
ENSDART00000056162 |
ftr13
|
finTRIM family, member 13 |
| chr12_-_14211067 | 0.11 |
ENSDART00000077903
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr2_+_32846602 | 0.11 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr23_-_4225606 | 0.11 |
ENSDART00000014152
ENSDART00000133341 |
aar2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr2_-_42173834 | 0.11 |
ENSDART00000098357
ENSDART00000144707 |
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr6_-_54433995 | 0.11 |
ENSDART00000017230
|
snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr13_-_42673978 | 0.11 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr5_+_54555567 | 0.11 |
ENSDART00000171159
|
anapc2
|
anaphase promoting complex subunit 2 |
| chr5_+_51833305 | 0.11 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr18_+_6638974 | 0.11 |
ENSDART00000162398
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr5_-_67292690 | 0.11 |
ENSDART00000062366
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr9_+_25853052 | 0.10 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr9_-_36924388 | 0.10 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr5_+_64856666 | 0.10 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr6_+_3334710 | 0.10 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr20_+_715739 | 0.10 |
ENSDART00000136768
|
myo6a
|
myosin VIa |
| chr25_+_8921425 | 0.10 |
ENSDART00000128591
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr24_+_41281265 | 0.10 |
ENSDART00000156920
ENSDART00000155151 |
nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr2_-_45510223 | 0.10 |
ENSDART00000113058
|
gpsm2
|
G protein signaling modulator 2 |
| chr18_-_26797723 | 0.10 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr13_-_33654931 | 0.10 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr3_+_36617024 | 0.10 |
ENSDART00000189957
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr17_+_8754020 | 0.10 |
ENSDART00000105322
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr18_-_39321484 | 0.09 |
ENSDART00000077694
|
leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr6_-_29159888 | 0.09 |
ENSDART00000110288
|
zbtb11
|
zinc finger and BTB domain containing 11 |
| chr3_-_24681404 | 0.09 |
ENSDART00000161612
|
BX569789.1
|
|
| chr3_-_34113838 | 0.09 |
ENSDART00000151216
|
ighv4-2
|
immunoglobulin heavy variable 4-2 |
| chr19_-_1871415 | 0.09 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr16_-_31824525 | 0.09 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr20_-_16223245 | 0.09 |
ENSDART00000014721
|
echdc2
|
enoyl CoA hydratase domain containing 2 |
| chr22_-_10752471 | 0.09 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr24_-_37640705 | 0.09 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr2_+_48303142 | 0.09 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr22_+_2959879 | 0.09 |
ENSDART00000185354
ENSDART00000063545 |
impact
|
impact RWD domain protein |
| chr12_-_9468618 | 0.08 |
ENSDART00000152737
ENSDART00000091519 |
pgap3
|
post-GPI attachment to proteins 3 |
| chr1_+_12335816 | 0.08 |
ENSDART00000067086
|
nansa
|
N-acetylneuraminic acid synthase a |
| chr20_-_48898560 | 0.08 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr13_-_18069421 | 0.08 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr10_+_43037064 | 0.08 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr20_-_20610812 | 0.08 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr4_-_12978925 | 0.08 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr2_-_56095275 | 0.08 |
ENSDART00000154701
|
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr19_-_34742440 | 0.08 |
ENSDART00000122625
ENSDART00000175621 |
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr3_+_32425202 | 0.08 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr14_-_5407118 | 0.08 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr25_+_17920668 | 0.08 |
ENSDART00000093358
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr7_-_32629458 | 0.08 |
ENSDART00000001376
|
arl14ep
|
ADP-ribosylation factor-like 14 effector protein |
| chr22_+_8753092 | 0.08 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr8_-_33154677 | 0.08 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr15_-_34933560 | 0.08 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr23_-_32404022 | 0.08 |
ENSDART00000156387
ENSDART00000155508 |
si:ch211-66i15.4
|
si:ch211-66i15.4 |
| chr8_-_17987547 | 0.08 |
ENSDART00000112699
ENSDART00000061747 |
fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr15_-_29114449 | 0.08 |
ENSDART00000145748
ENSDART00000109482 ENSDART00000179123 |
zgc:162698
|
zgc:162698 |
| chr24_+_17069420 | 0.08 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr6_-_54444929 | 0.07 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr14_-_31856819 | 0.07 |
ENSDART00000003345
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr18_+_27000850 | 0.07 |
ENSDART00000188906
ENSDART00000086131 |
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr14_-_30897177 | 0.07 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr4_-_1824836 | 0.07 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr14_+_712115 | 0.07 |
ENSDART00000157494
ENSDART00000166516 ENSDART00000082017 ENSDART00000122374 ENSDART00000170203 |
ints10
|
integrator complex subunit 10 |
| chr15_-_34668485 | 0.07 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr7_-_29356084 | 0.07 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr18_+_27001115 | 0.07 |
ENSDART00000133547
|
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr15_+_38299563 | 0.07 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr3_+_59880317 | 0.07 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr21_-_22892124 | 0.07 |
ENSDART00000065563
|
ccdc90b
|
coiled-coil domain containing 90B |
| chr14_-_43616572 | 0.07 |
ENSDART00000111189
|
gar1
|
GAR1 homolog, ribonucleoprotein |
| chr6_-_49526510 | 0.07 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr1_+_24557414 | 0.07 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr7_-_20103384 | 0.07 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr14_+_23687678 | 0.07 |
ENSDART00000002469
|
hspa4b
|
heat shock protein 4b |
| chr8_+_39770162 | 0.07 |
ENSDART00000190677
|
hps4
|
Hermansky-Pudlak syndrome 4 |
| chr16_+_54829293 | 0.07 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr6_-_2222707 | 0.07 |
ENSDART00000022179
|
prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr24_-_7826489 | 0.07 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr5_-_26795438 | 0.07 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr11_+_6374448 | 0.07 |
ENSDART00000135906
|
AL840638.2
|
|
| chr5_+_41477954 | 0.07 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr6_-_39080630 | 0.06 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr1_+_44582369 | 0.06 |
ENSDART00000003022
ENSDART00000137980 |
med19b
|
mediator complex subunit 19b |
| chr3_-_34136368 | 0.06 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr4_+_55055395 | 0.06 |
ENSDART00000159142
ENSDART00000099760 |
si:dkey-19c16.12
|
si:dkey-19c16.12 |
| chr24_+_3478871 | 0.06 |
ENSDART00000111491
ENSDART00000134598 ENSDART00000142407 |
wdr37
|
WD repeat domain 37 |
| chr5_+_61862199 | 0.06 |
ENSDART00000132452
|
sympk
|
symplekin |
| chr12_+_6465557 | 0.06 |
ENSDART00000066477
ENSDART00000122271 |
dkk1b
|
dickkopf WNT signaling pathway inhibitor 1b |
| chr3_-_34107685 | 0.06 |
ENSDART00000151130
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr5_+_47863153 | 0.06 |
ENSDART00000051518
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr9_-_41090048 | 0.06 |
ENSDART00000131681
ENSDART00000182552 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr17_+_27456804 | 0.06 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr15_+_25528290 | 0.06 |
ENSDART00000123143
|
npat
|
nuclear protein, ataxia-telangiectasia locus |
| chr18_+_3169579 | 0.06 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr8_-_21052371 | 0.06 |
ENSDART00000136561
|
si:dkeyp-82a1.6
|
si:dkeyp-82a1.6 |
| chr17_+_37253706 | 0.06 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr15_-_43284021 | 0.06 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr5_-_25084318 | 0.06 |
ENSDART00000089339
|
dph7
|
diphthamide biosynthesis 7 |
| chr25_-_34280080 | 0.06 |
ENSDART00000085251
|
gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr24_+_36392040 | 0.06 |
ENSDART00000136057
|
arf2b
|
ADP-ribosylation factor 2b |
| chr3_-_27066451 | 0.06 |
ENSDART00000156228
ENSDART00000156311 |
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr1_-_44048798 | 0.06 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr6_+_50451337 | 0.06 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr17_+_8754426 | 0.06 |
ENSDART00000185519
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr18_-_18584839 | 0.06 |
ENSDART00000159274
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr3_-_3413669 | 0.06 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr5_+_51833132 | 0.06 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr19_-_27006764 | 0.06 |
ENSDART00000089540
|
sacm1la
|
SAC1 like phosphatidylinositide phosphatase a |
| chr24_+_36392486 | 0.06 |
ENSDART00000007381
ENSDART00000135898 |
arf2b
|
ADP-ribosylation factor 2b |
| chr5_+_64397454 | 0.06 |
ENSDART00000015940
|
edf1
|
endothelial differentiation-related factor 1 |
| chr19_-_1855368 | 0.06 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr16_+_11029762 | 0.06 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr18_+_38755023 | 0.06 |
ENSDART00000010177
ENSDART00000193136 |
onecut1
|
one cut homeobox 1 |
| chr12_+_22823219 | 0.06 |
ENSDART00000153199
ENSDART00000192571 |
afap1
|
actin filament associated protein 1 |
| chr2_+_44571200 | 0.06 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr14_-_45558490 | 0.06 |
ENSDART00000165060
|
ints5
|
integrator complex subunit 5 |
| chr8_-_1266181 | 0.06 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPDEF
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0019264 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.1 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 | 0.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.1 | GO:0097201 | response to acidic pH(GO:0010447) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) |
| 0.0 | 0.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.0 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0051219 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |