Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
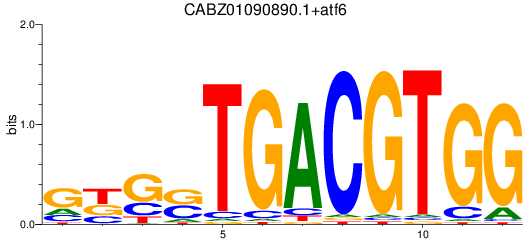
Results for CABZ01090890.1+atf6
Z-value: 0.72
Transcription factors associated with CABZ01090890.1+atf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf6
|
ENSDARG00000012656 | activating transcription factor 6 |
|
CABZ01090890.1
|
ENSDARG00000101369 | ENSDARG00000101369 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf6 | dr11_v1_chr20_-_33909872_33909872 | 0.74 | 1.5e-01 | Click! |
Activity profile of CABZ01090890.1+atf6 motif
Sorted Z-values of CABZ01090890.1+atf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_33767890 | 0.43 |
ENSDART00000193177
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr10_+_42374957 | 0.31 |
ENSDART00000147926
|
COX5B
|
zgc:86599 |
| chr13_+_33606739 | 0.28 |
ENSDART00000026464
|
cfl1l
|
cofilin 1 (non-muscle), like |
| chr1_+_38142715 | 0.24 |
ENSDART00000079928
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr4_-_8030583 | 0.24 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr10_+_42374770 | 0.24 |
ENSDART00000020000
|
COX5B
|
zgc:86599 |
| chr1_+_38142354 | 0.20 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr8_-_44298964 | 0.19 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr18_+_7553950 | 0.17 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr3_-_57762247 | 0.17 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr4_-_13921185 | 0.16 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr4_-_991043 | 0.16 |
ENSDART00000184706
|
naga
|
N-acetylgalactosaminidase, alpha |
| chr7_+_39166460 | 0.15 |
ENSDART00000052318
ENSDART00000146635 ENSDART00000173877 ENSDART00000173767 ENSDART00000173600 |
mdka
|
midkine a |
| chr4_-_25515154 | 0.14 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr11_-_3897067 | 0.14 |
ENSDART00000134858
|
rpn1
|
ribophorin I |
| chr6_+_55428924 | 0.14 |
ENSDART00000018270
|
ncoa5
|
nuclear receptor coactivator 5 |
| chr25_-_37084032 | 0.13 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr19_-_30404096 | 0.13 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr19_+_17400283 | 0.13 |
ENSDART00000127353
|
nr1d2b
|
nuclear receptor subfamily 1, group D, member 2b |
| chr6_+_13506841 | 0.13 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr19_-_30403922 | 0.13 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr17_-_24879003 | 0.13 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr3_+_36617024 | 0.12 |
ENSDART00000189957
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr3_+_22984098 | 0.12 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr12_-_13318944 | 0.12 |
ENSDART00000152201
ENSDART00000041394 |
emc9
|
ER membrane protein complex subunit 9 |
| chr5_-_9540641 | 0.12 |
ENSDART00000124384
ENSDART00000160079 |
gak
|
cyclin G associated kinase |
| chr17_-_14705039 | 0.12 |
ENSDART00000154281
ENSDART00000123550 |
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr2_+_10063747 | 0.11 |
ENSDART00000143876
ENSDART00000014088 ENSDART00000134554 |
cmpk
|
cytidylate kinase |
| chr15_-_25153352 | 0.11 |
ENSDART00000078095
ENSDART00000122184 |
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr13_+_17694845 | 0.11 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr10_+_16036246 | 0.11 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr15_+_47746176 | 0.11 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr14_+_23687678 | 0.11 |
ENSDART00000002469
|
hspa4b
|
heat shock protein 4b |
| chr18_-_40481028 | 0.11 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr22_+_24157807 | 0.10 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr9_+_8898835 | 0.10 |
ENSDART00000147820
|
naxd
|
NAD(P)HX dehydratase |
| chr18_+_8917766 | 0.10 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr9_+_21793565 | 0.10 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr10_+_16036573 | 0.10 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr1_+_10110203 | 0.10 |
ENSDART00000080576
ENSDART00000181437 |
lratb.1
|
lecithin retinol acyltransferase b, tandem duplicate 1 |
| chr16_-_54942532 | 0.10 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr3_+_36616713 | 0.09 |
ENSDART00000158284
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr10_-_15879569 | 0.09 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr21_-_40413191 | 0.09 |
ENSDART00000003221
|
nsrp1
|
nuclear speckle splicing regulatory protein 1 |
| chr21_-_31252131 | 0.09 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr14_-_4044545 | 0.09 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr13_-_45123051 | 0.09 |
ENSDART00000138644
|
runx3
|
runt-related transcription factor 3 |
| chr18_-_5248365 | 0.09 |
ENSDART00000082506
ENSDART00000082504 ENSDART00000097960 |
myef2
|
myelin expression factor 2 |
| chr1_+_26605065 | 0.09 |
ENSDART00000011645
|
coro2a
|
coronin, actin binding protein, 2A |
| chr7_-_44963154 | 0.09 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr15_-_3534705 | 0.09 |
ENSDART00000158150
|
cog6
|
component of oligomeric golgi complex 6 |
| chr23_+_7692042 | 0.09 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr12_+_46708920 | 0.09 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr6_-_29051773 | 0.09 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr5_-_30151815 | 0.09 |
ENSDART00000156048
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr10_-_8672820 | 0.08 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr16_+_1228073 | 0.08 |
ENSDART00000109645
|
LO017852.1
|
|
| chr12_+_2995887 | 0.08 |
ENSDART00000189499
ENSDART00000182597 ENSDART00000184918 ENSDART00000182292 |
lrrc45
|
leucine rich repeat containing 45 |
| chr7_+_19835569 | 0.08 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr8_-_2289264 | 0.08 |
ENSDART00000189397
|
plat
|
plasminogen activator, tissue |
| chr5_-_41804398 | 0.08 |
ENSDART00000134492
|
alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr4_+_12931763 | 0.08 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr25_+_245438 | 0.08 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr25_-_8513255 | 0.08 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr4_+_17642731 | 0.08 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr10_+_22510280 | 0.08 |
ENSDART00000109070
ENSDART00000182002 ENSDART00000192852 |
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr15_-_47937204 | 0.07 |
ENSDART00000154705
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr19_-_10324182 | 0.07 |
ENSDART00000151352
ENSDART00000151162 ENSDART00000023571 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr19_+_43297546 | 0.07 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr9_+_50038347 | 0.07 |
ENSDART00000164712
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr17_-_28797395 | 0.07 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr15_-_163586 | 0.07 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr9_+_890536 | 0.07 |
ENSDART00000139132
|
steap3
|
STEAP family member 3, metalloreductase |
| chr9_-_27396404 | 0.07 |
ENSDART00000136412
ENSDART00000101401 |
tex30
|
testis expressed 30 |
| chr3_-_34115886 | 0.07 |
ENSDART00000151531
|
ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr11_-_44945636 | 0.07 |
ENSDART00000157658
|
orc2
|
origin recognition complex, subunit 2 |
| chr7_+_33279108 | 0.07 |
ENSDART00000084530
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr9_+_8898024 | 0.06 |
ENSDART00000134954
ENSDART00000143671 ENSDART00000147098 ENSDART00000179969 ENSDART00000111214 ENSDART00000140232 ENSDART00000139687 ENSDART00000132443 ENSDART00000130208 |
naxd
|
NAD(P)HX dehydratase |
| chr4_-_25515513 | 0.06 |
ENSDART00000142276
ENSDART00000044043 |
rbm17
|
RNA binding motif protein 17 |
| chr23_+_33718602 | 0.06 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr10_-_28380919 | 0.06 |
ENSDART00000183409
ENSDART00000183105 ENSDART00000100207 ENSDART00000185392 ENSDART00000131220 |
btg3
|
B-cell translocation gene 3 |
| chr4_-_16883051 | 0.06 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr19_-_29294457 | 0.06 |
ENSDART00000130815
ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr15_-_35212462 | 0.06 |
ENSDART00000043960
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr2_-_11504347 | 0.06 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr22_+_2751887 | 0.06 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr3_-_26184018 | 0.06 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr22_-_10641873 | 0.06 |
ENSDART00000064772
|
cyb561d2
|
cytochrome b561 family, member D2 |
| chr22_+_1796057 | 0.06 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr25_+_37435720 | 0.06 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr4_+_357810 | 0.06 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr3_-_15889508 | 0.06 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr23_-_31266586 | 0.06 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr23_+_20431140 | 0.06 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr19_-_10324573 | 0.06 |
ENSDART00000171795
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr4_-_3145359 | 0.06 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr3_-_26183699 | 0.05 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr17_+_584369 | 0.05 |
ENSDART00000165143
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr17_-_49438873 | 0.05 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr18_+_20482369 | 0.05 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr12_+_33919502 | 0.05 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr14_-_45595711 | 0.05 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr19_+_4443285 | 0.05 |
ENSDART00000162683
|
trappc9
|
trafficking protein particle complex 9 |
| chr23_-_9859989 | 0.05 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr4_+_47257854 | 0.05 |
ENSDART00000173868
|
crestin
|
crestin |
| chr2_+_31833997 | 0.05 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr4_-_27099224 | 0.05 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr11_+_42482920 | 0.05 |
ENSDART00000160937
|
arf4a
|
ADP-ribosylation factor 4a |
| chr3_-_180860 | 0.05 |
ENSDART00000059956
ENSDART00000192506 |
kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr9_+_10014817 | 0.05 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr6_-_38930726 | 0.05 |
ENSDART00000154151
|
hdac7b
|
histone deacetylase 7b |
| chr14_-_215051 | 0.05 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr21_-_26677834 | 0.05 |
ENSDART00000077381
|
nxf1
|
nuclear RNA export factor 1 |
| chr3_+_431208 | 0.05 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr17_-_24866964 | 0.05 |
ENSDART00000190601
ENSDART00000192547 |
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr13_+_9896845 | 0.05 |
ENSDART00000169076
|
si:ch211-117n7.8
|
si:ch211-117n7.8 |
| chr23_+_21067684 | 0.05 |
ENSDART00000132066
|
kcnab2b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 b |
| chr24_-_35534273 | 0.05 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr6_-_28943056 | 0.05 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr5_-_19014589 | 0.05 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr22_-_21676364 | 0.05 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr22_+_1947494 | 0.05 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr16_-_45178430 | 0.05 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr1_-_18592068 | 0.05 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr2_-_38225388 | 0.05 |
ENSDART00000146485
ENSDART00000128043 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr19_+_31904836 | 0.04 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr15_+_17100697 | 0.04 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr3_-_22829710 | 0.04 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr18_-_34549721 | 0.04 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr16_-_52540056 | 0.04 |
ENSDART00000188304
|
CR293507.1
|
|
| chr22_+_32228882 | 0.04 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr11_+_42494531 | 0.04 |
ENSDART00000067604
|
arf4a
|
ADP-ribosylation factor 4a |
| chr20_-_1191910 | 0.04 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr20_+_26940178 | 0.04 |
ENSDART00000190888
|
cdca4
|
cell division cycle associated 4 |
| chr23_+_2703044 | 0.04 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr21_-_45382112 | 0.04 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr21_-_25295087 | 0.04 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr22_+_2254972 | 0.04 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr6_+_27275716 | 0.04 |
ENSDART00000156434
ENSDART00000114347 |
scly
|
selenocysteine lyase |
| chr10_+_2582254 | 0.04 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr14_-_11456724 | 0.04 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr19_-_1363759 | 0.04 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr22_+_2247645 | 0.04 |
ENSDART00000143366
ENSDART00000147852 |
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr17_+_21102301 | 0.04 |
ENSDART00000035432
|
entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr9_+_22780901 | 0.04 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr9_+_10014514 | 0.04 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr8_-_31919624 | 0.04 |
ENSDART00000085573
|
rgs7bpa
|
regulator of G protein signaling 7 binding protein a |
| chr22_+_2801464 | 0.04 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr4_+_33373100 | 0.04 |
ENSDART00000150417
|
znf1090
|
zinc finger protein 1090 |
| chr10_+_9595575 | 0.04 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr2_-_37465517 | 0.04 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr8_+_49065348 | 0.04 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr22_+_2937485 | 0.04 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr18_-_50524017 | 0.04 |
ENSDART00000150013
ENSDART00000149912 |
cd276
|
CD276 molecule |
| chr20_+_19212962 | 0.04 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr17_-_24866727 | 0.04 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr23_+_40655306 | 0.03 |
ENSDART00000141958
|
cdh24a
|
cadherin 24, type 2a |
| chr25_-_3034668 | 0.03 |
ENSDART00000053405
|
scamp2
|
secretory carrier membrane protein 2 |
| chr22_-_2937503 | 0.03 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr2_+_38225427 | 0.03 |
ENSDART00000141614
|
si:ch211-14a17.11
|
si:ch211-14a17.11 |
| chr11_-_22916641 | 0.03 |
ENSDART00000080201
ENSDART00000154813 |
mdm4
|
MDM4, p53 regulator |
| chr18_-_26894732 | 0.03 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr24_+_854877 | 0.03 |
ENSDART00000188408
|
piezo2a.2
|
piezo-type mechanosensitive ion channel component 2a, tandem duplicate 2 |
| chr5_+_3501859 | 0.03 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr2_-_3045861 | 0.03 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr23_-_2081554 | 0.03 |
ENSDART00000179805
|
ndnf
|
neuron-derived neurotrophic factor |
| chr20_-_51727860 | 0.03 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr2_+_51645164 | 0.03 |
ENSDART00000169600
|
abhd4
|
abhydrolase domain containing 4 |
| chr21_-_3613702 | 0.03 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr16_+_5678071 | 0.03 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr15_+_17100412 | 0.03 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr17_-_21617618 | 0.03 |
ENSDART00000109031
|
gpr26
|
G protein-coupled receptor 26 |
| chr9_+_54237100 | 0.03 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr19_+_40248697 | 0.03 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr13_-_25842074 | 0.03 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr1_-_36772147 | 0.03 |
ENSDART00000053369
|
prmt9
|
protein arginine methyltransferase 9 |
| chr3_+_5087787 | 0.02 |
ENSDART00000162787
|
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr6_+_59176470 | 0.02 |
ENSDART00000161720
|
shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr8_+_1843135 | 0.02 |
ENSDART00000141452
|
snap29
|
synaptosomal-associated protein 29 |
| chr7_-_33949246 | 0.02 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr15_-_37683768 | 0.02 |
ENSDART00000059609
|
si:dkey-117a8.3
|
si:dkey-117a8.3 |
| chr3_+_41731527 | 0.02 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr13_-_22699024 | 0.02 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr13_-_48431766 | 0.02 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr22_+_10713713 | 0.02 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr22_+_35275206 | 0.02 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr19_-_26736336 | 0.02 |
ENSDART00000109258
ENSDART00000182802 |
csnk2b
|
casein kinase 2, beta polypeptide |
| chr20_-_50014936 | 0.02 |
ENSDART00000148892
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr25_+_16080181 | 0.02 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr8_-_5220125 | 0.02 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr8_-_65189 | 0.02 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr22_-_32392252 | 0.02 |
ENSDART00000148681
ENSDART00000137945 |
pcbp4
|
poly(rC) binding protein 4 |
| chr13_+_36923052 | 0.02 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr3_-_55537096 | 0.02 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr15_+_41815703 | 0.02 |
ENSDART00000059508
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr5_-_27994679 | 0.02 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr3_-_47235997 | 0.02 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr18_+_50523870 | 0.02 |
ENSDART00000151593
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr15_-_37733238 | 0.02 |
ENSDART00000115178
|
si:dkey-42l23.7
|
si:dkey-42l23.7 |
| chr22_-_4989803 | 0.02 |
ENSDART00000181359
ENSDART00000125265 ENSDART00000028634 ENSDART00000183294 |
cherp
|
calcium homeostasis endoplasmic reticulum protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01090890.1+atf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.1 | 0.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.1 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |