Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
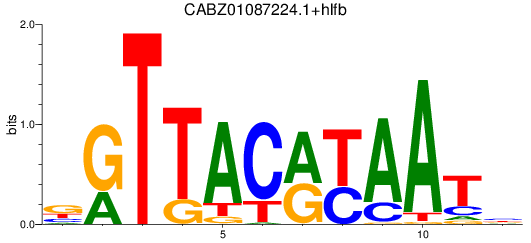
Results for CABZ01087224.1+hlfb
Z-value: 0.92
Transcription factors associated with CABZ01087224.1+hlfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hlfb
|
ENSDARG00000061011 | HLF transcription factor, PAR bZIP family member b |
|
CABZ01087224.1
|
ENSDARG00000111269 | ENSDARG00000111269 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CABZ01087224.1 | dr11_v1_chr3_+_11568523_11568523 | -0.33 | 5.9e-01 | Click! |
| hlfb | dr11_v1_chr12_+_32159272_32159272 | -0.22 | 7.2e-01 | Click! |
Activity profile of CABZ01087224.1+hlfb motif
Sorted Z-values of CABZ01087224.1+hlfb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_7735157 | 0.61 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr3_-_40275096 | 0.48 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr3_+_30921246 | 0.48 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr22_+_11775269 | 0.47 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr2_+_10134345 | 0.45 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr5_+_45677781 | 0.45 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr9_-_48736388 | 0.43 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr5_+_43782267 | 0.39 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr14_+_3507326 | 0.38 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr7_+_12950507 | 0.37 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr16_-_45917322 | 0.37 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr8_-_21103041 | 0.37 |
ENSDART00000171771
ENSDART00000131322 ENSDART00000137838 |
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr13_-_36034582 | 0.36 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr22_+_11756040 | 0.36 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr19_+_43715911 | 0.32 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr8_-_21103522 | 0.32 |
ENSDART00000100283
|
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr16_-_45917683 | 0.32 |
ENSDART00000184289
|
afp4
|
antifreeze protein type IV |
| chr22_-_17677947 | 0.31 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr5_+_22974019 | 0.31 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr5_-_69212184 | 0.30 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr25_-_13202208 | 0.29 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr8_-_50147948 | 0.29 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr16_-_29387215 | 0.29 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr4_+_5531583 | 0.28 |
ENSDART00000137500
ENSDART00000042080 |
si:dkey-14d8.6
|
si:dkey-14d8.6 |
| chr7_+_15736230 | 0.28 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr4_-_12725513 | 0.28 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr19_+_4990320 | 0.26 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr11_+_43419809 | 0.26 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr3_-_57744323 | 0.26 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr7_+_38380135 | 0.25 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr22_+_26443235 | 0.25 |
ENSDART00000044085
|
zgc:92480
|
zgc:92480 |
| chr19_-_17210760 | 0.25 |
ENSDART00000007906
|
stmn1a
|
stathmin 1a |
| chr22_+_24645325 | 0.24 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr14_+_32430982 | 0.24 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr17_+_26965351 | 0.24 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr7_-_12968689 | 0.24 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr24_+_38671054 | 0.24 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr13_-_25196758 | 0.24 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr24_-_26328721 | 0.24 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr13_+_25396896 | 0.23 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr10_-_15053507 | 0.23 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr22_+_10606863 | 0.22 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr5_+_6955900 | 0.22 |
ENSDART00000099417
|
CABZ01041962.1
|
|
| chr21_+_25625026 | 0.22 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr14_+_21699414 | 0.22 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr22_+_10606573 | 0.21 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr4_+_5196469 | 0.21 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr2_-_19234329 | 0.21 |
ENSDART00000161106
ENSDART00000160060 ENSDART00000174552 |
cdc20
|
cell division cycle 20 homolog |
| chr20_+_35857399 | 0.21 |
ENSDART00000102611
|
cd2ap
|
CD2-associated protein |
| chr22_-_817479 | 0.21 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr19_-_17210928 | 0.21 |
ENSDART00000164683
|
stmn1a
|
stathmin 1a |
| chr11_+_25504215 | 0.21 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr13_+_25397098 | 0.20 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr9_+_41218967 | 0.20 |
ENSDART00000000280
ENSDART00000145674 |
stat1b
|
signal transducer and activator of transcription 1b |
| chr6_-_39649504 | 0.20 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr5_+_25585869 | 0.20 |
ENSDART00000138060
|
si:dkey-229d2.7
|
si:dkey-229d2.7 |
| chr25_-_29415369 | 0.20 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr6_+_53288018 | 0.19 |
ENSDART00000170319
|
cysltr3
|
cysteinyl leukotriene receptor 3 |
| chr2_+_35595454 | 0.19 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr24_-_17023392 | 0.19 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr22_+_15336752 | 0.19 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr9_-_32300783 | 0.18 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr19_+_4990496 | 0.18 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr5_-_36948586 | 0.18 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr21_-_27443995 | 0.18 |
ENSDART00000003508
|
bfb
|
complement component bfb |
| chr12_+_22657925 | 0.18 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr1_-_10071422 | 0.18 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr9_-_9225980 | 0.18 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr6_-_10752937 | 0.18 |
ENSDART00000135093
|
ola1
|
Obg-like ATPase 1 |
| chr16_-_383664 | 0.18 |
ENSDART00000051693
|
irx4a
|
iroquois homeobox 4a |
| chr6_+_42475730 | 0.18 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr7_+_56577522 | 0.18 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr17_-_10838434 | 0.18 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr6_+_16468776 | 0.17 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr5_-_63509581 | 0.17 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr22_+_15315655 | 0.17 |
ENSDART00000141249
|
sult3st3
|
sulfotransferase family 3, cytosolic sulfotransferase 3 |
| chr15_+_19838458 | 0.17 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr10_-_23099809 | 0.17 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr9_-_44953349 | 0.17 |
ENSDART00000135156
|
vil1
|
villin 1 |
| chr20_-_25522911 | 0.17 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr16_-_51253925 | 0.17 |
ENSDART00000050644
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr1_-_58887610 | 0.17 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr3_-_26183699 | 0.17 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr18_+_25546227 | 0.16 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr2_+_44348473 | 0.16 |
ENSDART00000155166
ENSDART00000098146 |
zgc:152670
|
zgc:152670 |
| chr24_-_23784701 | 0.16 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr12_+_37401331 | 0.16 |
ENSDART00000125040
|
si:ch211-152f22.4
|
si:ch211-152f22.4 |
| chr21_-_33126697 | 0.16 |
ENSDART00000189293
ENSDART00000084559 |
CU019662.1
|
|
| chr6_+_36381709 | 0.16 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr5_+_29831235 | 0.16 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr4_+_77973876 | 0.16 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr6_-_26895750 | 0.16 |
ENSDART00000011863
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr5_-_11809404 | 0.15 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr15_-_23546452 | 0.15 |
ENSDART00000100669
|
si:dkey-182i3.11
|
si:dkey-182i3.11 |
| chr15_-_1765098 | 0.15 |
ENSDART00000149980
ENSDART00000093074 |
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr7_+_53156810 | 0.15 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr24_-_26310854 | 0.15 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr23_-_43486714 | 0.15 |
ENSDART00000169726
|
e2f1
|
E2F transcription factor 1 |
| chr24_+_26329018 | 0.14 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr12_+_19199735 | 0.14 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr21_+_1647990 | 0.14 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr24_+_26328787 | 0.14 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr19_+_43716607 | 0.14 |
ENSDART00000133199
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr8_+_4838114 | 0.14 |
ENSDART00000146667
|
es1
|
es1 protein |
| chr13_+_18533005 | 0.14 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr1_+_16600690 | 0.14 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr6_+_50451337 | 0.14 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr9_-_32300611 | 0.14 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr4_+_17353714 | 0.14 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr4_+_5341592 | 0.14 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr4_+_9478500 | 0.14 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr23_+_11285662 | 0.14 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr3_-_26184018 | 0.14 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr14_+_4151379 | 0.14 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr1_-_23274038 | 0.14 |
ENSDART00000181658
|
rfc1
|
replication factor C (activator 1) 1 |
| chr20_-_25533739 | 0.14 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr4_-_27099224 | 0.13 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr5_-_14521500 | 0.13 |
ENSDART00000176565
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr25_+_3104959 | 0.13 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr7_+_22792895 | 0.13 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr3_+_1107102 | 0.13 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr7_-_3429874 | 0.13 |
ENSDART00000132330
|
si:ch211-285c6.1
|
si:ch211-285c6.1 |
| chr14_+_21699129 | 0.13 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr13_-_10945288 | 0.13 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr23_-_18981595 | 0.13 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr17_-_6198823 | 0.13 |
ENSDART00000028407
ENSDART00000193636 |
ptk2ba
|
protein tyrosine kinase 2 beta, a |
| chr2_-_37277626 | 0.13 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr6_+_9130989 | 0.13 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr9_-_11550711 | 0.13 |
ENSDART00000093343
|
fev
|
FEV (ETS oncogene family) |
| chr19_-_30810328 | 0.13 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr25_+_36292465 | 0.13 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr16_+_23397785 | 0.13 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr6_-_1553314 | 0.13 |
ENSDART00000077209
|
tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chr19_+_14109348 | 0.13 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr7_+_46019780 | 0.12 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr19_-_3056235 | 0.12 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr10_+_38417512 | 0.12 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr6_+_29791164 | 0.12 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr12_-_13205854 | 0.12 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr12_-_13205572 | 0.12 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr8_+_12930216 | 0.12 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr13_+_10945337 | 0.12 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr14_+_16345003 | 0.12 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr5_-_36949476 | 0.12 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr25_-_13188678 | 0.12 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr19_+_14113886 | 0.12 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr9_+_13682133 | 0.12 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr9_-_12888082 | 0.11 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr2_-_55779927 | 0.11 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr11_+_12744575 | 0.11 |
ENSDART00000131059
ENSDART00000081335 ENSDART00000142481 |
rbb4l
|
retinoblastoma binding protein 4, like |
| chr7_+_67733759 | 0.11 |
ENSDART00000172015
|
cyb5b
|
cytochrome b5 type B |
| chr6_-_32411703 | 0.11 |
ENSDART00000151002
ENSDART00000078908 |
usp1
|
ubiquitin specific peptidase 1 |
| chr11_-_23459779 | 0.11 |
ENSDART00000183935
ENSDART00000184125 ENSDART00000193284 |
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr16_-_6849754 | 0.11 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr8_-_25846188 | 0.11 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr9_+_38292947 | 0.11 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr12_-_20584413 | 0.11 |
ENSDART00000170923
|
FP885542.2
|
|
| chr4_+_30785713 | 0.11 |
ENSDART00000165945
|
si:dkey-178j11.5
|
si:dkey-178j11.5 |
| chr8_-_42594380 | 0.11 |
ENSDART00000140126
ENSDART00000135238 ENSDART00000192764 |
dok2
|
docking protein 2 |
| chr18_-_16922905 | 0.11 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr16_+_23398369 | 0.11 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr9_+_30478768 | 0.11 |
ENSDART00000101097
|
acp6
|
acid phosphatase 6, lysophosphatidic |
| chr19_-_24757231 | 0.11 |
ENSDART00000128177
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr20_-_9760424 | 0.11 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr9_+_21306902 | 0.11 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr3_+_40284598 | 0.11 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr19_-_30811161 | 0.11 |
ENSDART00000103524
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr18_-_7448047 | 0.11 |
ENSDART00000193213
ENSDART00000131940 ENSDART00000186944 ENSDART00000052803 |
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr13_-_25774183 | 0.11 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr1_+_14253118 | 0.11 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr20_+_2039518 | 0.11 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr16_-_19568388 | 0.11 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr21_+_45841731 | 0.10 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr9_+_18023288 | 0.10 |
ENSDART00000098355
|
tnfsf11
|
TNF superfamily member 11 |
| chr15_+_12435975 | 0.10 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr22_-_10605045 | 0.10 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr6_-_48347724 | 0.10 |
ENSDART00000046973
|
capza1a
|
capping protein (actin filament) muscle Z-line, alpha 1a |
| chr5_-_2721686 | 0.10 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr22_+_1123873 | 0.10 |
ENSDART00000137708
|
si:ch1073-181h11.2
|
si:ch1073-181h11.2 |
| chr4_+_39766931 | 0.10 |
ENSDART00000138663
|
si:dkeyp-85d8.1
|
si:dkeyp-85d8.1 |
| chr8_-_37249813 | 0.10 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr8_+_247163 | 0.10 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr8_+_22472584 | 0.10 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr19_+_12649691 | 0.10 |
ENSDART00000192956
ENSDART00000088917 |
rnmt
|
RNA (guanine-7-) methyltransferase |
| chr11_-_23501467 | 0.10 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr13_-_42536642 | 0.10 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr10_+_39283985 | 0.10 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr14_+_8174828 | 0.10 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr10_+_23099890 | 0.10 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr19_+_16015881 | 0.10 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr24_-_26854032 | 0.10 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr25_-_26833100 | 0.10 |
ENSDART00000014052
|
neil1
|
nei-like DNA glycosylase 1 |
| chr5_-_30074332 | 0.10 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr7_+_46020508 | 0.10 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr9_-_11551608 | 0.10 |
ENSDART00000128629
|
fev
|
FEV (ETS oncogene family) |
| chr7_-_21887104 | 0.10 |
ENSDART00000019699
|
mettl3
|
methyltransferase like 3 |
| chr17_-_20236228 | 0.10 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr13_-_10620652 | 0.10 |
ENSDART00000135000
ENSDART00000191587 |
si:ch73-54n14.2
camkmt
|
si:ch73-54n14.2 calmodulin-lysine N-methyltransferase |
| chr25_+_34845115 | 0.10 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr7_+_22853788 | 0.09 |
ENSDART00000148502
|
clcn5b
|
chloride channel, voltage-sensitive 5b |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01087224.1+hlfb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 0.3 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.4 | GO:0031282 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.4 | GO:0051818 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.1 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.2 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.3 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.5 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of cyclase activity(GO:0031281) positive regulation of adenylate cyclase activity(GO:0045762) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.2 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.0 | 0.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.1 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0019474 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.2 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.0 | 0.8 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.1 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.0 | 0.1 | GO:0046436 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.2 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.1 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.4 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.3 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.0 | GO:0097268 | cytoophidium(GO:0097268) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.2 | 0.5 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.2 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.5 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.2 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.1 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.0 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |