Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
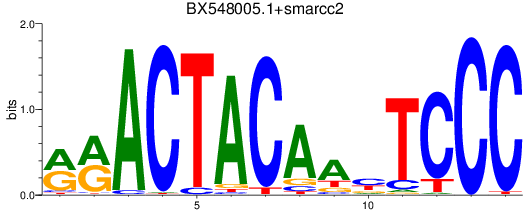
Results for BX548005.1+smarcc2
Z-value: 2.10
Transcription factors associated with BX548005.1+smarcc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smarcc2
|
ENSDARG00000077946 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
|
BX548005.1
|
ENSDARG00000110907 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smarcc2 | dr11_v1_chr6_+_39836474_39836474 | 0.93 | 2.4e-02 | Click! |
Activity profile of BX548005.1+smarcc2 motif
Sorted Z-values of BX548005.1+smarcc2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_65536095 | 1.87 |
ENSDART00000189898
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr10_+_36037977 | 1.79 |
ENSDART00000164678
|
katnal1
|
katanin p60 subunit A-like 1 |
| chr16_+_5774977 | 1.71 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr8_-_5155545 | 1.39 |
ENSDART00000056885
|
DPYSL2 (1 of many)
|
dihydropyrimidinase like 2 |
| chr16_-_34285106 | 1.38 |
ENSDART00000044235
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr25_+_10458990 | 1.34 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr18_+_45792035 | 1.34 |
ENSDART00000135045
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr6_-_13449556 | 1.29 |
ENSDART00000184566
ENSDART00000112883 ENSDART00000185377 |
fmnl2b
|
formin-like 2b |
| chr16_+_34523515 | 1.24 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr7_-_36358303 | 1.23 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr8_-_21268303 | 1.22 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr9_-_29985390 | 1.17 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr16_-_43971258 | 1.14 |
ENSDART00000141941
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr1_+_16073887 | 1.09 |
ENSDART00000160270
|
tusc3
|
tumor suppressor candidate 3 |
| chr12_+_7445595 | 1.08 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr11_-_10770053 | 1.08 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr21_-_43485351 | 1.00 |
ENSDART00000151597
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr16_+_34522977 | 0.99 |
ENSDART00000144069
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr24_+_25692802 | 0.98 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr6_+_58543336 | 0.94 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr13_+_33055548 | 0.94 |
ENSDART00000137315
|
rdh12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr19_-_5254699 | 0.94 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr21_-_23746916 | 0.92 |
ENSDART00000017229
|
ncam1a
|
neural cell adhesion molecule 1a |
| chr20_-_40319890 | 0.92 |
ENSDART00000075112
|
clvs2
|
clavesin 2 |
| chr15_-_2857961 | 0.89 |
ENSDART00000033263
|
ankrd49
|
ankyrin repeat domain 49 |
| chr8_-_39984593 | 0.86 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr16_-_34401412 | 0.85 |
ENSDART00000054020
|
hivep3b
|
human immunodeficiency virus type I enhancer binding protein 3b |
| chr25_-_5815834 | 0.85 |
ENSDART00000112853
|
appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr25_+_7229046 | 0.85 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr25_+_15273370 | 0.85 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr21_+_21791343 | 0.83 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr17_-_26604946 | 0.82 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr8_+_18624658 | 0.80 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr23_-_4019928 | 0.79 |
ENSDART00000021062
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr2_+_38608290 | 0.78 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr13_-_32626247 | 0.76 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr5_-_22082918 | 0.76 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr25_-_31763897 | 0.75 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr1_-_54947592 | 0.75 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr2_-_13254821 | 0.75 |
ENSDART00000022621
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr17_-_21418340 | 0.74 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr20_-_42100932 | 0.74 |
ENSDART00000191930
|
slc35f1
|
solute carrier family 35, member F1 |
| chr2_+_53357953 | 0.73 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr21_+_21791799 | 0.72 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr19_-_1023051 | 0.72 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr2_-_44720551 | 0.71 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr8_+_1148876 | 0.71 |
ENSDART00000148651
|
avp
|
arginine vasopressin |
| chr4_-_27398385 | 0.71 |
ENSDART00000142117
ENSDART00000150553 ENSDART00000182746 |
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr17_-_8692722 | 0.70 |
ENSDART00000148931
ENSDART00000192891 |
ctbp2a
|
C-terminal binding protein 2a |
| chr18_+_2189211 | 0.70 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr20_-_45661049 | 0.69 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr5_-_23675222 | 0.69 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr22_-_38516922 | 0.68 |
ENSDART00000151257
ENSDART00000151785 |
klc4
|
kinesin light chain 4 |
| chr4_+_11479705 | 0.68 |
ENSDART00000019458
ENSDART00000150587 ENSDART00000139370 ENSDART00000135826 |
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr12_+_48480632 | 0.67 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr4_-_5092335 | 0.67 |
ENSDART00000130453
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr6_-_27057702 | 0.67 |
ENSDART00000149363
|
stk25a
|
serine/threonine kinase 25a |
| chr3_+_14339728 | 0.67 |
ENSDART00000184127
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr17_+_25414033 | 0.66 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr17_-_12336987 | 0.66 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr21_-_43065657 | 0.66 |
ENSDART00000180076
ENSDART00000190751 |
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr7_-_55292116 | 0.66 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr15_-_14375452 | 0.65 |
ENSDART00000160675
ENSDART00000164028 ENSDART00000171642 |
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr8_+_21629941 | 0.65 |
ENSDART00000140145
|
ajap1
|
adherens junctions associated protein 1 |
| chr5_-_12407194 | 0.65 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr17_-_945883 | 0.63 |
ENSDART00000158837
|
dync1h1
|
dynein, cytoplasmic 1, heavy chain 1 |
| chr7_-_36358735 | 0.63 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr6_+_48664275 | 0.62 |
ENSDART00000161184
|
FAM19A3
|
si:dkey-238f9.1 |
| chr5_-_41142467 | 0.62 |
ENSDART00000129415
|
zfr
|
zinc finger RNA binding protein |
| chr6_-_13188667 | 0.62 |
ENSDART00000191654
|
adam23a
|
ADAM metallopeptidase domain 23a |
| chr25_+_3347461 | 0.62 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr23_-_12158685 | 0.61 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr23_-_33654889 | 0.61 |
ENSDART00000146180
|
csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr21_-_30284404 | 0.60 |
ENSDART00000066363
|
zgc:175066
|
zgc:175066 |
| chr9_-_23217196 | 0.60 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr5_-_31716713 | 0.60 |
ENSDART00000131443
|
dpm2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr10_-_37337579 | 0.59 |
ENSDART00000147705
|
omga
|
oligodendrocyte myelin glycoprotein a |
| chr19_-_32150078 | 0.59 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr16_-_30655980 | 0.58 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr16_+_23495600 | 0.58 |
ENSDART00000021092
|
snx27b
|
sorting nexin family member 27b |
| chr1_+_49878000 | 0.58 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr14_-_30876708 | 0.58 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr20_+_38525567 | 0.58 |
ENSDART00000147787
|
znf512
|
zinc finger protein 512 |
| chr25_-_8916913 | 0.58 |
ENSDART00000104629
ENSDART00000131748 |
furinb
|
furin (paired basic amino acid cleaving enzyme) b |
| chr8_-_40276035 | 0.57 |
ENSDART00000185301
ENSDART00000187610 ENSDART00000053158 ENSDART00000162020 |
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr2_-_13254594 | 0.57 |
ENSDART00000155671
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr17_-_16324565 | 0.57 |
ENSDART00000030835
|
hmbox1a
|
homeobox containing 1a |
| chr5_-_41142768 | 0.56 |
ENSDART00000074789
|
zfr
|
zinc finger RNA binding protein |
| chr1_+_6172786 | 0.56 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr17_-_24439672 | 0.56 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr16_+_25137483 | 0.55 |
ENSDART00000155666
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr2_-_32352946 | 0.55 |
ENSDART00000144870
ENSDART00000077151 |
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr17_-_12764360 | 0.55 |
ENSDART00000003418
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr24_-_33276139 | 0.54 |
ENSDART00000128943
|
nrbp2b
|
nuclear receptor binding protein 2b |
| chr14_+_45306073 | 0.54 |
ENSDART00000034606
ENSDART00000173301 |
sfxn5b
|
sideroflexin 5b |
| chr13_-_49169545 | 0.54 |
ENSDART00000192076
|
tsnax
|
translin-associated factor X |
| chr12_-_31103187 | 0.52 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr14_-_493029 | 0.52 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr22_-_29689485 | 0.52 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr1_+_29281764 | 0.52 |
ENSDART00000112106
|
fam155a
|
family with sequence similarity 155, member A |
| chr21_+_28496194 | 0.51 |
ENSDART00000138236
|
cox8a
|
cytochrome c oxidase subunit VIIIA (ubiquitous) |
| chr6_-_53282572 | 0.51 |
ENSDART00000172615
ENSDART00000165036 |
rbm5
|
RNA binding motif protein 5 |
| chr11_-_23025949 | 0.51 |
ENSDART00000184859
ENSDART00000167818 ENSDART00000046122 ENSDART00000193120 |
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr19_+_177455 | 0.51 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr12_-_48467733 | 0.51 |
ENSDART00000153126
ENSDART00000152895 ENSDART00000014190 |
sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr14_-_30876299 | 0.51 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr13_+_11073282 | 0.50 |
ENSDART00000114911
ENSDART00000182462 ENSDART00000148100 ENSDART00000137816 ENSDART00000134888 ENSDART00000131320 |
sdccag8
|
serologically defined colon cancer antigen 8 |
| chr17_+_9017775 | 0.49 |
ENSDART00000186901
ENSDART00000185407 |
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr24_+_15899365 | 0.49 |
ENSDART00000185965
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr15_-_11341635 | 0.49 |
ENSDART00000055220
|
rab30
|
RAB30, member RAS oncogene family |
| chr11_-_18283886 | 0.48 |
ENSDART00000019248
|
stimate
|
STIM activating enhance |
| chr20_+_4060839 | 0.48 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr23_-_4019699 | 0.48 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr15_+_15516612 | 0.48 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr24_-_9960290 | 0.47 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr25_-_20258508 | 0.47 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr19_+_2546775 | 0.47 |
ENSDART00000148527
ENSDART00000097528 |
sp4
|
sp4 transcription factor |
| chr15_+_22867174 | 0.46 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr13_-_11072964 | 0.46 |
ENSDART00000135989
|
cep170aa
|
centrosomal protein 170Aa |
| chr7_-_29043075 | 0.46 |
ENSDART00000052342
|
thap11
|
THAP domain containing 11 |
| chr22_+_22888 | 0.46 |
ENSDART00000082471
|
mfap2
|
microfibril associated protein 2 |
| chr3_+_23488652 | 0.45 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr20_-_33790003 | 0.45 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr5_-_12687357 | 0.45 |
ENSDART00000193206
|
aifm3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr7_+_20876303 | 0.45 |
ENSDART00000173495
ENSDART00000164172 |
gigyf1a
|
GRB10 interacting GYF protein 1a |
| chr2_+_5841108 | 0.45 |
ENSDART00000136180
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr12_+_22680115 | 0.44 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr15_-_9419472 | 0.44 |
ENSDART00000122117
|
sacs
|
sacsin molecular chaperone |
| chr22_+_10129788 | 0.44 |
ENSDART00000045752
|
tbc1d20
|
TBC1 domain family, member 20 |
| chr5_-_62988463 | 0.43 |
ENSDART00000047143
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr6_-_50730749 | 0.43 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr2_-_52841762 | 0.42 |
ENSDART00000114682
|
ralbp1
|
ralA binding protein 1 |
| chr11_+_37049347 | 0.42 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr25_+_18889332 | 0.42 |
ENSDART00000017043
|
cax2
|
cation/H+ exchanger protein 2 |
| chr23_+_31405497 | 0.42 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr4_+_25912654 | 0.41 |
ENSDART00000109508
ENSDART00000134218 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr12_+_48390715 | 0.41 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr15_+_21744680 | 0.41 |
ENSDART00000016860
|
ppp2r1bb
|
protein phosphatase 2, regulatory subunit A, beta b |
| chr21_-_38031038 | 0.40 |
ENSDART00000179483
ENSDART00000076238 |
rbm41
|
RNA binding motif protein 41 |
| chr15_+_8767650 | 0.40 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr24_+_19591893 | 0.40 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr11_+_8152872 | 0.40 |
ENSDART00000091638
ENSDART00000138057 ENSDART00000166379 |
samd13
|
sterile alpha motif domain containing 13 |
| chr10_+_37182626 | 0.40 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr24_+_39576071 | 0.40 |
ENSDART00000147137
|
si:dkey-161j23.6
|
si:dkey-161j23.6 |
| chr19_+_15521997 | 0.39 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr20_-_16849306 | 0.39 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr10_+_21650828 | 0.39 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr24_+_25032340 | 0.39 |
ENSDART00000005845
|
mtmr6
|
myotubularin related protein 6 |
| chr6_+_58522557 | 0.39 |
ENSDART00000128062
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr8_+_16726386 | 0.39 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr3_+_24060454 | 0.39 |
ENSDART00000143088
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr16_-_27161410 | 0.39 |
ENSDART00000177503
|
LO017771.1
|
|
| chr5_-_26795438 | 0.39 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr18_-_27316599 | 0.39 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr21_+_8427059 | 0.39 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr17_-_26604549 | 0.39 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr4_+_25912308 | 0.38 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr6_-_16394528 | 0.38 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr9_-_23242684 | 0.38 |
ENSDART00000053282
ENSDART00000179770 |
ccnt2a
|
cyclin T2a |
| chr6_-_10780698 | 0.38 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr4_+_15605844 | 0.37 |
ENSDART00000101619
ENSDART00000021384 |
exoc4
|
exocyst complex component 4 |
| chr16_-_41131578 | 0.37 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr11_+_23957440 | 0.36 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr13_-_15929402 | 0.36 |
ENSDART00000090273
|
ttl
|
tubulin tyrosine ligase |
| chr11_+_37049105 | 0.36 |
ENSDART00000155408
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr15_-_29556757 | 0.36 |
ENSDART00000060049
|
hspa13
|
heat shock protein 70 family, member 13 |
| chr10_+_8091144 | 0.35 |
ENSDART00000143848
ENSDART00000075485 |
sub1a
|
SUB1 homolog, transcriptional regulator a |
| chr8_-_46321889 | 0.35 |
ENSDART00000075189
ENSDART00000122801 |
mtor
|
mechanistic target of rapamycin kinase |
| chr5_+_37379825 | 0.35 |
ENSDART00000171826
|
klhl13
|
kelch-like family member 13 |
| chr11_+_27133560 | 0.35 |
ENSDART00000158411
|
hdac11
|
histone deacetylase 11 |
| chr21_-_11054876 | 0.35 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr19_+_40379771 | 0.35 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr16_-_2390931 | 0.34 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr13_+_2617555 | 0.34 |
ENSDART00000162208
|
plpp4
|
phospholipid phosphatase 4 |
| chr1_-_22803147 | 0.33 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr5_+_41143563 | 0.33 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr19_-_6083761 | 0.33 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr7_+_15618127 | 0.32 |
ENSDART00000080160
|
lrrc28
|
leucine rich repeat containing 28 |
| chr7_+_74134010 | 0.32 |
ENSDART00000164874
|
cldnd1a
|
claudin domain containing 1a |
| chr3_-_26904774 | 0.32 |
ENSDART00000103690
|
clec16a
|
C-type lectin domain containing 16A |
| chr18_+_9641210 | 0.32 |
ENSDART00000182024
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr6_+_4229360 | 0.32 |
ENSDART00000191347
ENSDART00000130642 |
FO082877.1
|
|
| chr11_-_44999858 | 0.31 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr23_-_14216506 | 0.31 |
ENSDART00000019620
|
ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr1_+_40237276 | 0.31 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr17_+_43659940 | 0.31 |
ENSDART00000145738
ENSDART00000075619 |
adob
|
2-aminoethanethiol (cysteamine) dioxygenase b |
| chr8_-_31384607 | 0.31 |
ENSDART00000164134
ENSDART00000024872 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr23_+_43668756 | 0.31 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr22_-_20342260 | 0.30 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr5_-_22019061 | 0.30 |
ENSDART00000113066
|
amer1
|
APC membrane recruitment protein 1 |
| chr25_-_21716326 | 0.30 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr4_+_11483335 | 0.30 |
ENSDART00000184978
ENSDART00000140954 |
asb13a.2
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 2 |
| chr19_+_18627100 | 0.30 |
ENSDART00000167245
|
vps52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr16_+_25107344 | 0.29 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr2_-_1468258 | 0.29 |
ENSDART00000114431
|
gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr24_+_15897717 | 0.29 |
ENSDART00000105956
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr25_-_4733008 | 0.29 |
ENSDART00000191884
|
drd4a
|
dopamine receptor D4a |
| chr19_-_17385548 | 0.28 |
ENSDART00000162383
|
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr8_-_36618073 | 0.28 |
ENSDART00000047912
|
gpkow
|
G patch domain and KOW motifs |
| chr1_+_52130213 | 0.28 |
ENSDART00000018817
|
rnf11a
|
ring finger protein 11a |
Network of associatons between targets according to the STRING database.
First level regulatory network of BX548005.1+smarcc2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0042245 | RNA repair(GO:0042245) |
| 0.3 | 1.3 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.3 | 0.9 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.2 | 1.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 1.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.9 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.7 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.2 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.3 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 1.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.4 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.3 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.4 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.7 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.7 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.4 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.6 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.8 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 1.2 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 0.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.6 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 1.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.5 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0045161 | neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 2.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 1.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 1.4 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.0 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.6 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.7 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.6 | GO:0034111 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.3 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.0 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.4 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.3 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 0.6 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.2 | 0.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 0.7 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.5 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 2.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.4 | 1.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 1.0 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 1.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.6 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.7 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.6 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.3 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.4 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.4 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.5 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.2 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 2.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.8 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.0 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.0 | 0.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |