Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
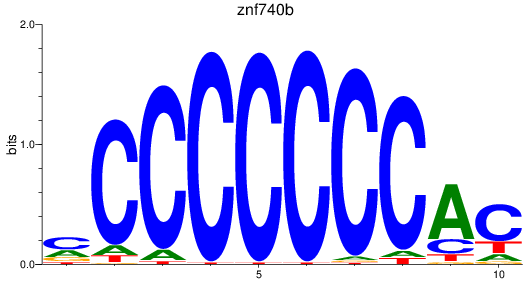
Results for znf740b
Z-value: 1.43
Transcription factors associated with znf740b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf740b
|
ENSDARG00000061174 | zinc finger protein 740b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf740b | dr11_v1_chr23_-_36418059_36418059 | -0.92 | 3.9e-04 | Click! |
Activity profile of znf740b motif
Sorted Z-values of znf740b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_24191928 | 3.79 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr14_-_9522364 | 3.71 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr21_-_26495700 | 3.00 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr3_-_61162750 | 2.92 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr12_-_16764751 | 2.90 |
ENSDART00000113862
|
zgc:174154
|
zgc:174154 |
| chr25_+_35502552 | 2.70 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr5_-_14326959 | 2.65 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr11_-_3334248 | 2.52 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr12_-_16941319 | 2.50 |
ENSDART00000109968
|
zgc:174855
|
zgc:174855 |
| chr25_+_1732838 | 2.48 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr23_-_524780 | 2.43 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr23_+_36101185 | 2.20 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr20_+_26538137 | 2.07 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr25_+_37366698 | 2.04 |
ENSDART00000165400
ENSDART00000192589 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr7_-_2039060 | 2.02 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr8_+_6954984 | 1.95 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr13_-_45523026 | 1.89 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr14_+_33458294 | 1.86 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr6_+_52804267 | 1.79 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr23_-_10175898 | 1.74 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr3_-_31804481 | 1.70 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr21_-_43117327 | 1.69 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr3_+_23703704 | 1.59 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr2_-_16565690 | 1.57 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr2_+_22694382 | 1.56 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr23_+_32499916 | 1.52 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr1_-_38709551 | 1.49 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr25_+_13191391 | 1.48 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr13_-_31435137 | 1.47 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr21_+_30351256 | 1.47 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr23_-_17470146 | 1.43 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr7_-_69636502 | 1.42 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr8_+_7311478 | 1.40 |
ENSDART00000133358
|
ssuh2rs1
|
ssu-2 homolog, related sequence 1 |
| chr20_+_20672163 | 1.40 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr12_+_13244149 | 1.38 |
ENSDART00000186984
ENSDART00000105896 |
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr17_+_52822422 | 1.37 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr18_+_44649804 | 1.35 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr20_+_25340814 | 1.33 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr2_+_43469241 | 1.32 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr2_-_23004286 | 1.31 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr20_-_35246150 | 1.26 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr12_-_4540564 | 1.26 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr4_-_5597167 | 1.25 |
ENSDART00000132431
|
vegfab
|
vascular endothelial growth factor Ab |
| chr3_+_40809011 | 1.22 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr12_+_42436328 | 1.20 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr23_+_42434348 | 1.20 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr7_-_48665305 | 1.19 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr11_-_97817 | 1.18 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr22_+_28446557 | 1.18 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr2_-_44183613 | 1.14 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr2_-_44183451 | 1.14 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr22_+_28446365 | 1.13 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr1_-_42778510 | 1.12 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr19_+_5315987 | 1.11 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr10_+_8968203 | 1.10 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr9_-_18877597 | 1.09 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr15_-_5815006 | 1.09 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr5_-_16351306 | 1.08 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr25_+_25464630 | 1.08 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr18_+_45990394 | 1.08 |
ENSDART00000024068
|
mmp23bb
|
matrix metallopeptidase 23bb |
| chr4_+_76619791 | 1.06 |
ENSDART00000184042
|
ms4a17a.8
|
membrane-spanning 4-domains, subfamily A, member 17A.8 |
| chr25_-_11088839 | 1.06 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr11_-_497854 | 1.05 |
ENSDART00000104520
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr7_+_71335815 | 1.05 |
ENSDART00000102756
|
sod3a
|
superoxide dismutase 3, extracellular a |
| chr19_+_42983613 | 1.04 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr10_-_43540196 | 1.03 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr21_+_7823146 | 1.03 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr23_+_44580254 | 1.02 |
ENSDART00000185069
|
pfn1
|
profilin 1 |
| chr11_+_38280454 | 1.01 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr21_+_19648814 | 1.01 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr16_-_8280885 | 0.99 |
ENSDART00000129068
|
entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr7_-_27686021 | 0.99 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr5_-_40510397 | 0.98 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr3_-_1190132 | 0.96 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr3_-_16289826 | 0.95 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr1_+_39553040 | 0.93 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_42779075 | 0.93 |
ENSDART00000133917
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr9_+_5862040 | 0.93 |
ENSDART00000129117
|
pdzk1
|
PDZ domain containing 1 |
| chr8_-_23776399 | 0.93 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr5_-_71838520 | 0.92 |
ENSDART00000174396
|
CU927890.1
|
|
| chr2_+_23790748 | 0.92 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr10_-_43113731 | 0.92 |
ENSDART00000138099
|
tmem167a
|
transmembrane protein 167A |
| chr10_-_43294933 | 0.91 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr3_+_50312422 | 0.90 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr23_-_19715557 | 0.90 |
ENSDART00000143764
|
rpl10
|
ribosomal protein L10 |
| chr25_+_13191615 | 0.88 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr24_-_37877743 | 0.85 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr7_-_35126374 | 0.84 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr16_+_22587661 | 0.84 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr5_+_30699260 | 0.83 |
ENSDART00000012888
|
atp5l
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g |
| chr17_-_36988455 | 0.82 |
ENSDART00000187180
ENSDART00000126823 |
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr18_+_21122818 | 0.81 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr16_+_10918252 | 0.80 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr10_+_10210455 | 0.78 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr5_-_41841675 | 0.76 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr12_-_20584413 | 0.76 |
ENSDART00000170923
|
FP885542.2
|
|
| chr22_-_15957041 | 0.76 |
ENSDART00000149236
ENSDART00000187500 ENSDART00000176304 ENSDART00000080047 ENSDART00000190068 |
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr2_-_6051836 | 0.76 |
ENSDART00000092479
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr24_+_41931585 | 0.76 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr9_-_32753535 | 0.73 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr11_-_18705303 | 0.72 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr24_-_21674950 | 0.72 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr13_+_23677949 | 0.70 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr8_+_8291492 | 0.70 |
ENSDART00000151314
|
srpk3
|
SRSF protein kinase 3 |
| chr23_+_36083529 | 0.69 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr16_+_10963602 | 0.69 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr18_-_34549721 | 0.69 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr19_+_2279051 | 0.68 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr20_-_16972351 | 0.68 |
ENSDART00000148312
ENSDART00000186702 |
si:ch73-74h11.1
|
si:ch73-74h11.1 |
| chr7_+_38249858 | 0.67 |
ENSDART00000150158
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr22_+_35068046 | 0.67 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr17_-_20849879 | 0.66 |
ENSDART00000088100
ENSDART00000149630 |
ank3b
|
ankyrin 3b |
| chr3_+_62126981 | 0.65 |
ENSDART00000060527
|
drc3
|
dynein regulatory complex subunit 3 |
| chr19_+_48117995 | 0.65 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr23_+_5977965 | 0.64 |
ENSDART00000115403
ENSDART00000183147 |
NAV1 (1 of many)
|
neuron navigator 1 |
| chr15_-_5742531 | 0.63 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr19_+_35799384 | 0.63 |
ENSDART00000076023
|
angpt2b
|
angiopoietin 2b |
| chr3_-_33417826 | 0.62 |
ENSDART00000084284
|
abi3a
|
ABI family, member 3a |
| chr21_+_25660613 | 0.62 |
ENSDART00000134017
|
si:dkey-17e16.15
|
si:dkey-17e16.15 |
| chr23_+_6232895 | 0.61 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr1_+_27070760 | 0.61 |
ENSDART00000186790
|
bnc2
|
basonuclin 2 |
| chr3_-_26204867 | 0.61 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr25_-_37501371 | 0.61 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr9_+_12959760 | 0.60 |
ENSDART00000125961
|
bmpr2b
|
bone morphogenetic protein receptor, type II b (serine/threonine kinase) |
| chr4_+_12612145 | 0.60 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr10_-_31563049 | 0.60 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr18_+_35742838 | 0.59 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr5_+_32162684 | 0.59 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr8_-_23578660 | 0.59 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr10_+_2742499 | 0.59 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr6_+_54239625 | 0.58 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr17_-_12389259 | 0.58 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr7_+_61480296 | 0.58 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr7_-_57949117 | 0.58 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr1_-_51474974 | 0.57 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr21_+_19635486 | 0.57 |
ENSDART00000185736
|
fgf10a
|
fibroblast growth factor 10a |
| chr2_+_28889936 | 0.56 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr10_-_35542071 | 0.56 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr2_+_47623202 | 0.55 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr9_-_31278048 | 0.55 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr20_-_37629084 | 0.55 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr23_-_41651759 | 0.54 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr23_+_20931030 | 0.54 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr1_-_44638058 | 0.54 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr1_-_52498146 | 0.54 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr24_-_28243186 | 0.54 |
ENSDART00000105691
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr12_+_22258962 | 0.54 |
ENSDART00000131175
|
wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr20_-_42100932 | 0.54 |
ENSDART00000191930
|
slc35f1
|
solute carrier family 35, member F1 |
| chr16_-_41439659 | 0.53 |
ENSDART00000191624
|
cpne4a
|
copine IVa |
| chr24_-_21808816 | 0.52 |
ENSDART00000025621
ENSDART00000130446 |
flt1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) |
| chr7_+_46261199 | 0.52 |
ENSDART00000170390
ENSDART00000183227 |
znf536
|
zinc finger protein 536 |
| chr24_-_40860603 | 0.51 |
ENSDART00000188032
|
CU633479.7
|
|
| chr25_-_18470695 | 0.51 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr3_+_21189766 | 0.50 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr4_+_11311315 | 0.50 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr1_+_31112436 | 0.50 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr21_-_11820379 | 0.49 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr8_+_36803415 | 0.48 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr12_-_7854216 | 0.48 |
ENSDART00000149594
|
ank3b
|
ankyrin 3b |
| chr16_-_31435020 | 0.47 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr11_-_4235811 | 0.47 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr21_+_5635420 | 0.47 |
ENSDART00000168158
|
shroom3
|
shroom family member 3 |
| chr11_+_6650966 | 0.47 |
ENSDART00000131236
|
si:dkey-246j7.1
|
si:dkey-246j7.1 |
| chr13_-_638485 | 0.46 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr7_+_36552725 | 0.46 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_68022631 | 0.46 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr6_+_38381957 | 0.45 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr21_-_37889727 | 0.45 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr1_+_53842344 | 0.45 |
ENSDART00000114506
|
crfb17
|
cytokine receptor family member B17 |
| chr21_+_42930558 | 0.44 |
ENSDART00000135234
|
stk32a
|
serine/threonine kinase 32A |
| chr9_+_8942258 | 0.44 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr5_-_70625697 | 0.43 |
ENSDART00000179538
ENSDART00000190540 |
CABZ01075572.1
|
|
| chr13_-_25198025 | 0.43 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr6_+_23712911 | 0.43 |
ENSDART00000167795
|
zgc:158654
|
zgc:158654 |
| chr3_-_52614747 | 0.43 |
ENSDART00000154365
|
trim35-13
|
tripartite motif containing 35-13 |
| chr21_+_5589923 | 0.43 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr12_+_5530247 | 0.42 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr19_-_25519310 | 0.42 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr12_+_24561832 | 0.42 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr6_-_37460178 | 0.41 |
ENSDART00000153500
|
si:dkey-66a8.7
|
si:dkey-66a8.7 |
| chr16_-_23800484 | 0.41 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr2_-_26527730 | 0.41 |
ENSDART00000138693
|
si:ch211-106k21.5
|
si:ch211-106k21.5 |
| chr1_+_51475094 | 0.41 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr15_-_35246742 | 0.40 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr15_+_3284416 | 0.40 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr8_+_8936912 | 0.38 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr18_-_16181952 | 0.38 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr4_+_34417403 | 0.37 |
ENSDART00000186032
ENSDART00000167909 |
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr4_+_3980247 | 0.37 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr6_+_15373153 | 0.36 |
ENSDART00000155865
|
tmtops2a
|
teleost multiple tissue opsin 2a |
| chr3_+_6469754 | 0.35 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr15_-_12229874 | 0.35 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr24_-_37877554 | 0.34 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr19_-_25519612 | 0.34 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr19_-_46957968 | 0.34 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr17_-_22062364 | 0.33 |
ENSDART00000114470
|
ttbk1b
|
tau tubulin kinase 1b |
| chr10_+_42159471 | 0.32 |
ENSDART00000122160
|
CR925773.1
|
|
| chr23_+_9353552 | 0.32 |
ENSDART00000163298
|
BX511246.1
|
|
| chr20_-_28642061 | 0.31 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr9_+_7358749 | 0.31 |
ENSDART00000081660
|
ihha
|
Indian hedgehog homolog a |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf740b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.5 | 2.6 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.4 | 1.6 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 1.5 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 1.0 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.3 | 0.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 1.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.9 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 1.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 2.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 0.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 1.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 1.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 1.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.8 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.2 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.5 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 1.0 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.7 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.6 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.4 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.7 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 1.6 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 2.0 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.3 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.2 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.1 | 2.7 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.4 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.4 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.5 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 1.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.0 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.8 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.1 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 2.2 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.6 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 1.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.8 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 1.5 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.1 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.6 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 1.2 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.1 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 1.3 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0060612 | adipose tissue development(GO:0060612) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 2.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 4.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 7.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 5.1 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 2.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 2.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 1.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.3 | 1.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 1.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 2.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.4 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.0 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.6 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.0 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 4.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.0 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |