Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
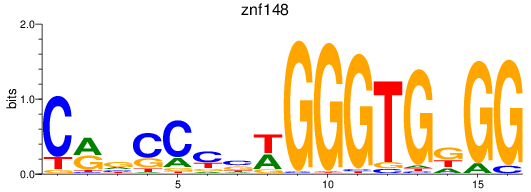
Results for znf148
Z-value: 0.81
Transcription factors associated with znf148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf148
|
ENSDARG00000055106 | zinc finger protein 148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf148 | dr11_v1_chr9_+_38624478_38624533 | 0.86 | 2.8e-03 | Click! |
Activity profile of znf148 motif
Sorted Z-values of znf148 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_233143 | 2.26 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr3_-_31804481 | 2.06 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr6_-_8736766 | 1.92 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr9_-_7539297 | 1.90 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr24_-_40667800 | 1.75 |
ENSDART00000169315
|
smyhc1
|
slow myosin heavy chain 1 |
| chr11_+_41981959 | 1.69 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr24_-_40668208 | 1.59 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr15_-_29388012 | 1.39 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr6_+_36459897 | 1.35 |
ENSDART00000104248
ENSDART00000145753 |
lrrc15
|
leucine rich repeat containing 15 |
| chr11_-_18705303 | 1.30 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr6_-_9581949 | 1.25 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr9_-_30274412 | 1.21 |
ENSDART00000089526
|
otc
|
ornithine carbamoyltransferase |
| chr11_+_44579865 | 1.17 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr3_-_37758487 | 1.16 |
ENSDART00000150938
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr4_-_11751037 | 1.11 |
ENSDART00000102301
|
podxl
|
podocalyxin-like |
| chr11_+_6116503 | 1.10 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr20_+_52554352 | 1.06 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr14_+_7939398 | 0.94 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr22_-_11438627 | 0.92 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr14_+_7939216 | 0.89 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr6_+_28877306 | 0.89 |
ENSDART00000065137
ENSDART00000123189 ENSDART00000065135 ENSDART00000181512 ENSDART00000130799 |
tp63
|
tumor protein p63 |
| chr14_+_18781082 | 0.78 |
ENSDART00000108793
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr20_+_17739923 | 0.78 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr1_-_411331 | 0.75 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr16_+_5678071 | 0.74 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr17_-_52521002 | 0.73 |
ENSDART00000114931
|
prox2
|
prospero homeobox 2 |
| chr24_-_3419998 | 0.73 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr12_-_41618844 | 0.72 |
ENSDART00000160054
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr6_+_12865137 | 0.71 |
ENSDART00000090065
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr10_+_38512270 | 0.68 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr4_+_37406676 | 0.67 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr20_+_305035 | 0.66 |
ENSDART00000104807
|
si:dkey-119m7.4
|
si:dkey-119m7.4 |
| chr6_+_50451337 | 0.65 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr8_+_31821396 | 0.62 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr13_-_36535128 | 0.60 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr5_-_26118855 | 0.55 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr9_-_6927587 | 0.54 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr10_+_22724059 | 0.53 |
ENSDART00000136123
|
kdm6bb
|
lysine (K)-specific demethylase 6B, b |
| chr16_+_40560622 | 0.51 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr13_-_40302710 | 0.51 |
ENSDART00000144360
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr25_+_13791627 | 0.49 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr3_-_1938588 | 0.48 |
ENSDART00000013001
ENSDART00000186405 |
zgc:152753
|
zgc:152753 |
| chr21_-_41838284 | 0.46 |
ENSDART00000141067
|
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr25_-_16554757 | 0.46 |
ENSDART00000154480
|
si:ch211-266k8.6
|
si:ch211-266k8.6 |
| chr10_+_1638876 | 0.45 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr10_+_31951338 | 0.45 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr3_-_61592417 | 0.44 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr13_+_12045475 | 0.43 |
ENSDART00000163053
ENSDART00000160812 ENSDART00000158244 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr13_+_23214100 | 0.42 |
ENSDART00000163393
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr22_-_607812 | 0.42 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr14_-_26177156 | 0.39 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr18_+_33725576 | 0.38 |
ENSDART00000146816
|
si:dkey-145c18.5
|
si:dkey-145c18.5 |
| chr16_+_11029762 | 0.38 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr12_-_41619257 | 0.35 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr13_+_12045758 | 0.35 |
ENSDART00000079398
ENSDART00000165467 ENSDART00000165880 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr6_+_4084739 | 0.34 |
ENSDART00000087661
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr20_+_7180298 | 0.34 |
ENSDART00000143026
|
si:dkeyp-51f12.2
|
si:dkeyp-51f12.2 |
| chr24_+_1294176 | 0.31 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr2_+_6181383 | 0.30 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr23_+_28648864 | 0.30 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr2_-_12243213 | 0.30 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr6_-_52156427 | 0.30 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr11_-_7029897 | 0.28 |
ENSDART00000077406
ENSDART00000184582 ENSDART00000180369 |
cdh27
|
cadherin 27 |
| chr5_+_4564233 | 0.28 |
ENSDART00000193435
|
CABZ01058647.1
|
|
| chr5_+_483965 | 0.28 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr22_-_38360205 | 0.27 |
ENSDART00000162055
|
mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr17_-_36818176 | 0.26 |
ENSDART00000061762
|
myo6b
|
myosin VIb |
| chr5_+_41793001 | 0.26 |
ENSDART00000136439
ENSDART00000190477 ENSDART00000192289 |
bcl7a
|
BCL tumor suppressor 7A |
| chr1_+_27052530 | 0.25 |
ENSDART00000184065
|
bnc2
|
basonuclin 2 |
| chr1_+_27053010 | 0.25 |
ENSDART00000161169
|
bnc2
|
basonuclin 2 |
| chr12_-_4532066 | 0.24 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr18_-_14337065 | 0.21 |
ENSDART00000135703
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr22_+_30035719 | 0.21 |
ENSDART00000192238
|
AL845330.1
|
|
| chr10_-_15053507 | 0.21 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr13_-_42579437 | 0.20 |
ENSDART00000135172
|
fam83ha
|
family with sequence similarity 83, member Ha |
| chr24_-_21490628 | 0.18 |
ENSDART00000181546
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr4_+_11464255 | 0.18 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr16_-_21181128 | 0.16 |
ENSDART00000133403
|
pde11al
|
phosphodiesterase 11a, like |
| chr5_+_1933131 | 0.15 |
ENSDART00000061693
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr2_+_10147029 | 0.15 |
ENSDART00000139064
ENSDART00000053426 ENSDART00000153678 |
pfn2l
|
profilin 2 like |
| chr21_-_11632403 | 0.13 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr8_-_1956632 | 0.13 |
ENSDART00000131602
|
si:dkey-178e17.1
|
si:dkey-178e17.1 |
| chr20_-_48172556 | 0.12 |
ENSDART00000097888
|
CABZ01059099.1
|
|
| chr3_-_31924643 | 0.11 |
ENSDART00000122589
|
rnf113a
|
ring finger protein 113A |
| chr9_-_11549379 | 0.10 |
ENSDART00000187074
|
fev
|
FEV (ETS oncogene family) |
| chr18_+_33373811 | 0.10 |
ENSDART00000156665
ENSDART00000048182 |
v2rh32
|
vomeronasal 2 receptor, h32 |
| chr14_+_46291022 | 0.10 |
ENSDART00000074099
|
cabp2b
|
calcium binding protein 2b |
| chr2_-_12242695 | 0.09 |
ENSDART00000158175
|
gpr158b
|
G protein-coupled receptor 158b |
| chr5_-_25621386 | 0.09 |
ENSDART00000051565
|
cyp1d1
|
cytochrome P450, family 1, subfamily D, polypeptide 1 |
| chr4_-_6459863 | 0.09 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr1_-_9227804 | 0.08 |
ENSDART00000190360
|
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr3_-_2591942 | 0.08 |
ENSDART00000127971
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr5_-_21030934 | 0.07 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr19_-_11015238 | 0.07 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr9_+_30274637 | 0.07 |
ENSDART00000079095
|
rpgra
|
retinitis pigmentosa GTPase regulator a |
| chr2_-_24907741 | 0.06 |
ENSDART00000155013
|
si:dkey-149i17.11
|
si:dkey-149i17.11 |
| chr6_-_141564 | 0.06 |
ENSDART00000151245
ENSDART00000063876 |
s1pr5b
|
sphingosine-1-phosphate receptor 5b |
| chr3_+_40796110 | 0.04 |
ENSDART00000014729
|
arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr7_-_1918971 | 0.02 |
ENSDART00000181759
|
CABZ01040626.1
|
|
| chr7_-_71837213 | 0.01 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr4_-_72468562 | 0.01 |
ENSDART00000181890
|
CR788316.1
|
|
| chr23_-_351935 | 0.00 |
ENSDART00000140986
|
uhrf1bp1
|
UHRF1 binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf148
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.5 | 1.9 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.4 | 1.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.3 | 1.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.3 | 0.8 | GO:0090248 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.1 | 0.9 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.7 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 1.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 2.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.3 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.3 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 | 1.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.9 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.8 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 1.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 1.7 | GO:0001756 | somitogenesis(GO:0001756) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 4.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |