Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
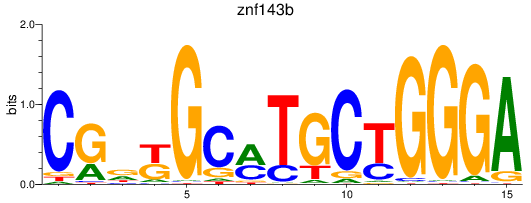
Results for znf143b
Z-value: 0.77
Transcription factors associated with znf143b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf143b
|
ENSDARG00000041581 | zinc finger protein 143b |
|
znf143b
|
ENSDARG00000111018 | zinc finger protein 143b |
|
znf143b
|
ENSDARG00000112408 | zinc finger protein 143b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf143b | dr11_v1_chr18_-_16937008_16937109 | 0.90 | 1.1e-03 | Click! |
Activity profile of znf143b motif
Sorted Z-values of znf143b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_25103965 | 1.12 |
ENSDART00000192561
ENSDART00000173721 |
si:dkey-23i12.9
|
si:dkey-23i12.9 |
| chr21_-_43636595 | 1.05 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr5_-_32383475 | 0.95 |
ENSDART00000141294
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr24_-_26622423 | 0.85 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr16_+_31511739 | 0.79 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr11_+_27134116 | 0.76 |
ENSDART00000129736
|
hdac11
|
histone deacetylase 11 |
| chr17_+_23470967 | 0.75 |
ENSDART00000104718
ENSDART00000154716 |
kif20ba
|
kinesin family member 20Ba |
| chr10_+_37182626 | 0.74 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr3_-_3448095 | 0.70 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr9_-_55946377 | 0.67 |
ENSDART00000161536
ENSDART00000169432 |
SH3RF3
|
si:ch211-124n19.2 |
| chr17_+_46387086 | 0.66 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr9_+_33158191 | 0.64 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr20_-_15161502 | 0.63 |
ENSDART00000187072
|
plpp6
|
phospholipid phosphatase 6 |
| chr4_-_75047192 | 0.63 |
ENSDART00000193576
|
large1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr17_-_25861787 | 0.63 |
ENSDART00000182503
|
BX000364.5
|
|
| chr17_+_22760126 | 0.60 |
ENSDART00000151999
ENSDART00000190442 |
ttc27
|
tetratricopeptide repeat domain 27 |
| chr15_+_5116179 | 0.60 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr15_+_44184367 | 0.60 |
ENSDART00000162918
ENSDART00000110060 |
zgc:165514
|
zgc:165514 |
| chr12_-_1034383 | 0.59 |
ENSDART00000152455
ENSDART00000152346 |
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr22_-_10891213 | 0.58 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr11_+_5880562 | 0.58 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr25_+_8925934 | 0.56 |
ENSDART00000073914
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr13_+_15816573 | 0.55 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr11_-_24532988 | 0.55 |
ENSDART00000067078
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr2_-_34138400 | 0.54 |
ENSDART00000056667
|
cenpl
|
centromere protein L |
| chr8_-_51524446 | 0.54 |
ENSDART00000167394
ENSDART00000147742 |
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr11_-_2222440 | 0.54 |
ENSDART00000162167
|
smug1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr18_+_18000887 | 0.54 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr2_-_34138064 | 0.54 |
ENSDART00000133381
|
cenpl
|
centromere protein L |
| chr22_+_26600834 | 0.54 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr5_-_68641353 | 0.53 |
ENSDART00000048432
|
dlg4a
|
discs, large homolog 4a (Drosophila) |
| chr19_+_31532043 | 0.53 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr19_+_40069524 | 0.53 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr15_+_25452092 | 0.52 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr14_+_32926385 | 0.52 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr3_+_18050667 | 0.52 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr18_-_41164277 | 0.52 |
ENSDART00000187766
ENSDART00000185375 |
CABZ01005876.1
|
|
| chr14_+_41409697 | 0.51 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr22_+_797105 | 0.50 |
ENSDART00000128549
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr9_-_8297344 | 0.50 |
ENSDART00000180945
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr2_-_37537887 | 0.50 |
ENSDART00000143496
ENSDART00000025841 |
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr16_+_42471455 | 0.49 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr2_+_42072231 | 0.49 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr8_+_19977712 | 0.48 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr16_-_8132742 | 0.48 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr3_+_35608385 | 0.48 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr19_-_6631900 | 0.48 |
ENSDART00000144571
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr7_+_67748939 | 0.48 |
ENSDART00000162978
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr4_+_14981854 | 0.47 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr20_-_15161669 | 0.46 |
ENSDART00000080333
ENSDART00000063882 |
plpp6
|
phospholipid phosphatase 6 |
| chr19_-_33996277 | 0.46 |
ENSDART00000159384
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr15_-_1622468 | 0.46 |
ENSDART00000149008
ENSDART00000034456 |
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr22_+_12366516 | 0.46 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr9_-_1639884 | 0.46 |
ENSDART00000062850
ENSDART00000150947 |
agps
|
alkylglycerone phosphate synthase |
| chr25_-_13871118 | 0.44 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr18_+_18000695 | 0.43 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr6_-_17999776 | 0.43 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr2_-_57110477 | 0.43 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr20_+_43691208 | 0.43 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr2_+_47718605 | 0.43 |
ENSDART00000189180
ENSDART00000148824 |
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr23_-_45568816 | 0.42 |
ENSDART00000076086
|
cyp17a2
|
cytochrome P450, family 17, subfamily A, polypeptide 2 |
| chr3_-_31057624 | 0.42 |
ENSDART00000152901
|
armc5
|
armadillo repeat containing 5 |
| chr10_-_27009413 | 0.42 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr19_-_34970312 | 0.41 |
ENSDART00000102896
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr2_+_42072689 | 0.41 |
ENSDART00000134203
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr21_-_16399778 | 0.41 |
ENSDART00000136435
|
unc5da
|
unc-5 netrin receptor Da |
| chr24_+_36840652 | 0.41 |
ENSDART00000088168
|
CABZ01055347.1
|
|
| chr3_+_29179329 | 0.41 |
ENSDART00000085216
ENSDART00000190136 |
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr14_+_32852388 | 0.40 |
ENSDART00000166351
|
nkrf
|
NFKB repressing factor |
| chr12_+_48581588 | 0.40 |
ENSDART00000114996
|
CABZ01078415.1
|
|
| chr1_+_53279861 | 0.40 |
ENSDART00000035713
|
rnf150a
|
ring finger protein 150a |
| chr21_+_7605803 | 0.40 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr7_-_11605185 | 0.39 |
ENSDART00000169291
ENSDART00000113904 |
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr22_-_506522 | 0.38 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr10_-_8079737 | 0.38 |
ENSDART00000059014
ENSDART00000179549 |
zgc:173443
si:ch211-251f6.6
|
zgc:173443 si:ch211-251f6.6 |
| chr8_+_21254192 | 0.38 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr22_-_5918670 | 0.38 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr5_+_34549845 | 0.37 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr3_-_40202607 | 0.36 |
ENSDART00000074757
|
znf598
|
zinc finger protein 598 |
| chr19_-_7358184 | 0.36 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr6_+_38773376 | 0.36 |
ENSDART00000078128
ENSDART00000184053 |
ube3a
|
ubiquitin protein ligase E3A |
| chr2_-_13691834 | 0.35 |
ENSDART00000186570
|
CABZ01044235.1
|
|
| chr13_-_51846224 | 0.35 |
ENSDART00000184663
|
LT631684.2
|
|
| chr24_+_39129316 | 0.35 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr23_+_43870886 | 0.35 |
ENSDART00000102658
ENSDART00000149088 |
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr17_+_44463230 | 0.35 |
ENSDART00000130311
|
naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr4_-_8058214 | 0.35 |
ENSDART00000132228
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr15_-_15968883 | 0.34 |
ENSDART00000166583
ENSDART00000154042 |
synrg
|
synergin, gamma |
| chr24_+_37449893 | 0.33 |
ENSDART00000164405
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr16_+_46401756 | 0.33 |
ENSDART00000147370
ENSDART00000144000 |
rpz2
|
rapunzel 2 |
| chr25_-_20258508 | 0.33 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr10_-_35186310 | 0.33 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr21_-_23046606 | 0.33 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr17_+_37253706 | 0.33 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr15_-_766015 | 0.32 |
ENSDART00000190648
|
si:dkey-7i4.15
|
si:dkey-7i4.15 |
| chr2_+_31437547 | 0.32 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr14_-_21661015 | 0.32 |
ENSDART00000189403
ENSDART00000172442 ENSDART00000181913 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr2_+_44571200 | 0.32 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr21_+_21791343 | 0.31 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr23_+_33947874 | 0.31 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr16_-_34212912 | 0.31 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr5_-_13353810 | 0.31 |
ENSDART00000146136
ENSDART00000099602 ENSDART00000004539 |
ube2l3b
|
ubiquitin-conjugating enzyme E2L 3b |
| chr19_+_41701660 | 0.31 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr23_-_270847 | 0.31 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr15_-_9257136 | 0.31 |
ENSDART00000187901
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr21_+_7131970 | 0.31 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr1_+_55095314 | 0.31 |
ENSDART00000132727
|
aftpha
|
aftiphilin a |
| chr19_+_43715911 | 0.30 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr24_+_39576071 | 0.30 |
ENSDART00000147137
|
si:dkey-161j23.6
|
si:dkey-161j23.6 |
| chr7_-_62003831 | 0.29 |
ENSDART00000113585
ENSDART00000062704 |
plaa
|
phospholipase A2-activating protein |
| chr15_-_18200358 | 0.29 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr19_-_1023051 | 0.29 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr5_-_66160415 | 0.28 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr13_-_46687097 | 0.28 |
ENSDART00000169106
ENSDART00000158202 |
CABZ01078449.1
|
|
| chr20_-_42534504 | 0.28 |
ENSDART00000132310
|
rfx6
|
regulatory factor X, 6 |
| chr5_+_34549365 | 0.28 |
ENSDART00000009500
|
aif1l
|
allograft inflammatory factor 1-like |
| chr2_+_53357953 | 0.27 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr5_-_15321451 | 0.27 |
ENSDART00000139203
|
txnrd2.1
|
thioredoxin reductase 2, tandem duplicate 1 |
| chr19_-_12648122 | 0.27 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr6_+_12482599 | 0.27 |
ENSDART00000090316
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr5_-_8907819 | 0.26 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr15_+_24676905 | 0.26 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr25_+_418932 | 0.26 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr20_+_14810886 | 0.26 |
ENSDART00000010550
|
pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr18_-_15329454 | 0.26 |
ENSDART00000191903
ENSDART00000019818 |
ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr10_+_42678520 | 0.26 |
ENSDART00000182496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr12_+_532019 | 0.25 |
ENSDART00000152272
|
si:dkey-94l16.4
|
si:dkey-94l16.4 |
| chr18_+_8231138 | 0.25 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr22_+_38919265 | 0.25 |
ENSDART00000148051
|
senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr4_+_288633 | 0.25 |
ENSDART00000183304
|
FO834823.1
|
|
| chr21_+_21791799 | 0.25 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr25_-_37227491 | 0.25 |
ENSDART00000156647
|
glg1a
|
golgi glycoprotein 1a |
| chr4_+_1619584 | 0.25 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr8_+_24299321 | 0.24 |
ENSDART00000112046
ENSDART00000183558 ENSDART00000183796 ENSDART00000181395 ENSDART00000184555 ENSDART00000172531 ENSDART00000191663 ENSDART00000184103 ENSDART00000183676 |
ZNF335
|
si:ch211-269m15.3 |
| chr4_+_11483335 | 0.24 |
ENSDART00000184978
ENSDART00000140954 |
asb13a.2
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 2 |
| chr20_+_44498056 | 0.24 |
ENSDART00000023763
|
wdcp
|
WD repeat and coiled coil containing |
| chr17_-_14780578 | 0.24 |
ENSDART00000154690
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr3_+_55031685 | 0.24 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr8_+_247163 | 0.23 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr10_+_589501 | 0.23 |
ENSDART00000188415
|
LO018557.1
|
|
| chr21_-_2565825 | 0.23 |
ENSDART00000169633
|
col4a3bpb
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein b |
| chr25_+_8662469 | 0.23 |
ENSDART00000154680
ENSDART00000188568 |
man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr15_-_9419472 | 0.23 |
ENSDART00000122117
|
sacs
|
sacsin molecular chaperone |
| chr3_+_32477782 | 0.22 |
ENSDART00000151261
|
scaf1
|
SR-related CTD-associated factor 1 |
| chr6_-_50730749 | 0.22 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr20_-_9428021 | 0.22 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b |
| chr1_-_49950643 | 0.22 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr17_+_8754426 | 0.22 |
ENSDART00000185519
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr9_-_11676491 | 0.22 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr10_-_36618674 | 0.22 |
ENSDART00000135302
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr2_+_11670270 | 0.22 |
ENSDART00000100524
|
ftr01
|
finTRIM family, member 1 |
| chr21_+_4320769 | 0.21 |
ENSDART00000168553
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr1_-_2334254 | 0.21 |
ENSDART00000144500
|
si:ch211-235f1.3
|
si:ch211-235f1.3 |
| chr8_-_43158486 | 0.21 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr24_-_31092711 | 0.21 |
ENSDART00000168398
|
hccsb
|
holocytochrome c synthase b |
| chr15_-_47193564 | 0.20 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr20_+_46699021 | 0.20 |
ENSDART00000167398
ENSDART00000029894 |
wdr26b
|
WD repeat domain 26b |
| chr23_+_37185247 | 0.20 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr15_+_33843596 | 0.20 |
ENSDART00000169037
|
inpp5ka
|
inositol polyphosphate-5-phosphatase Ka |
| chr3_+_62219158 | 0.19 |
ENSDART00000162354
|
BX470259.5
|
|
| chr7_+_67749251 | 0.18 |
ENSDART00000167562
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr5_-_62988463 | 0.18 |
ENSDART00000047143
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr23_+_7692042 | 0.18 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr13_-_27620815 | 0.18 |
ENSDART00000139904
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr18_-_8380090 | 0.17 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr10_+_7719796 | 0.17 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr11_+_3246059 | 0.17 |
ENSDART00000161529
|
timeless
|
timeless circadian clock |
| chr9_+_27876146 | 0.17 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr16_-_39900665 | 0.17 |
ENSDART00000136719
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr8_+_46610020 | 0.17 |
ENSDART00000082509
|
tas1r2.2
|
taste receptor, type 1, member 2, tandem duplicate 2 |
| chr16_+_19637384 | 0.17 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr4_-_9780931 | 0.16 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr20_-_14321359 | 0.16 |
ENSDART00000145104
|
dlgap2b
|
discs, large (Drosophila) homolog-associated protein 2b |
| chr3_-_30861177 | 0.16 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr11_-_1935883 | 0.16 |
ENSDART00000172885
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr3_-_34816893 | 0.16 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr24_+_19863246 | 0.15 |
ENSDART00000165242
|
CU638714.1
|
|
| chr16_-_5143124 | 0.15 |
ENSDART00000131876
ENSDART00000060630 |
ttk
|
ttk protein kinase |
| chr1_-_55196103 | 0.15 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr14_-_30876708 | 0.15 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr2_+_34138376 | 0.14 |
ENSDART00000137170
|
dars2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr9_-_29579908 | 0.14 |
ENSDART00000140876
|
cenpj
|
centromere protein J |
| chr18_-_11060548 | 0.14 |
ENSDART00000146692
|
tspan9a
|
tetraspanin 9a |
| chr25_-_25646806 | 0.14 |
ENSDART00000089066
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr18_+_36806545 | 0.14 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr21_+_13785975 | 0.14 |
ENSDART00000134682
|
slc31a2
|
solute carrier family 31 (copper transporter), member 2 |
| chr7_-_8344243 | 0.14 |
ENSDART00000149510
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr9_-_29985390 | 0.13 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr3_-_33573252 | 0.13 |
ENSDART00000191339
ENSDART00000184612 ENSDART00000187232 ENSDART00000190671 |
CR847537.1
|
|
| chr20_-_46158987 | 0.13 |
ENSDART00000132026
|
taar10a
|
trace amine-associated receptor 10a |
| chr1_-_21563040 | 0.13 |
ENSDART00000049572
|
ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr5_+_20148671 | 0.13 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr16_-_39267185 | 0.12 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr5_+_63767376 | 0.12 |
ENSDART00000138898
|
rgs3b
|
regulator of G protein signaling 3b |
| chr13_+_30506781 | 0.12 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr6_-_37468971 | 0.11 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr9_+_27354653 | 0.11 |
ENSDART00000134134
|
tlr20.2
|
toll-like receptor 20, tandem duplicate 2 |
| chr23_-_45318760 | 0.11 |
ENSDART00000166883
|
ccdc171
|
coiled-coil domain containing 171 |
| chr25_-_21609645 | 0.11 |
ENSDART00000087817
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr11_+_19060079 | 0.11 |
ENSDART00000158123
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf143b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.2 | 0.9 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 0.7 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 0.5 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.2 | 1.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 0.5 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.3 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.4 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.4 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.4 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.6 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.2 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.0 | 0.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.3 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.3 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) |
| 0.0 | 0.7 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.5 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.2 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.4 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.5 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.6 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.6 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |