Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
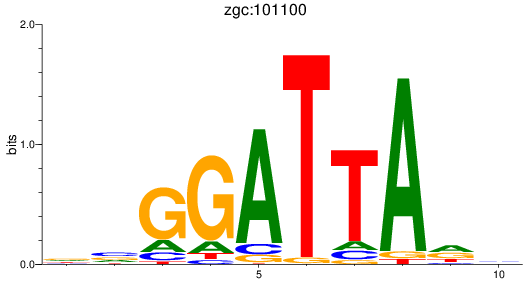
Results for zgc:101100
Z-value: 0.43
Transcription factors associated with zgc:101100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zgc
|
ENSDARG00000021959 | 101100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zgc:101100 | dr11_v1_chr8_+_20488322_20488322 | -0.28 | 4.7e-01 | Click! |
Activity profile of zgc:101100 motif
Sorted Z-values of zgc:101100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_54077740 | 0.61 |
ENSDART00000027000
|
rho
|
rhodopsin |
| chr6_+_41186320 | 0.44 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr11_-_15090118 | 0.38 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr11_-_15090564 | 0.36 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr14_+_32838110 | 0.33 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr5_+_11407504 | 0.28 |
ENSDART00000186723
ENSDART00000018047 |
zgc:112294
|
zgc:112294 |
| chr8_+_23142946 | 0.26 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr17_-_51202339 | 0.24 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr14_+_32837914 | 0.24 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr7_-_13882988 | 0.23 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr9_+_13714379 | 0.22 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr3_-_28828242 | 0.21 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr9_-_29544720 | 0.16 |
ENSDART00000130317
|
arhgap20
|
Rho GTPase activating protein 20 |
| chr10_+_17026870 | 0.15 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr2_-_5356686 | 0.15 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr22_-_910926 | 0.14 |
ENSDART00000180075
|
FP016205.1
|
|
| chr24_-_7777389 | 0.14 |
ENSDART00000138541
|
rpgrip1
|
RPGR interacting protein 1 |
| chr20_-_29475172 | 0.14 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr2_-_30784198 | 0.14 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr25_+_3326885 | 0.13 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr12_+_41991635 | 0.13 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr22_+_34701848 | 0.13 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr3_-_49566364 | 0.12 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr8_-_14121634 | 0.12 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr25_+_19041329 | 0.12 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr16_-_17188294 | 0.11 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr20_-_18736281 | 0.10 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr19_+_1097393 | 0.10 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr7_-_57933736 | 0.10 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr7_-_13884610 | 0.09 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr8_+_2656231 | 0.09 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr16_+_32059785 | 0.09 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr1_+_47585700 | 0.09 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr3_+_34919810 | 0.09 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr17_-_15149192 | 0.08 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr9_+_13685921 | 0.08 |
ENSDART00000145775
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr14_-_2933185 | 0.07 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr1_+_50613868 | 0.07 |
ENSDART00000111114
|
HERC5
|
si:ch73-190m4.1 |
| chr10_-_36738619 | 0.07 |
ENSDART00000093000
ENSDART00000157179 |
PLEKHB1
|
si:ch211-176g13.7 |
| chr7_+_66884291 | 0.06 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr3_+_34220194 | 0.06 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr20_-_25436389 | 0.06 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr10_-_26274094 | 0.06 |
ENSDART00000108798
|
dchs1b
|
dachsous cadherin-related 1b |
| chr2_+_30786773 | 0.06 |
ENSDART00000019029
ENSDART00000145681 |
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr7_+_66884570 | 0.06 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr9_-_12682848 | 0.05 |
ENSDART00000138360
|
myo10l3
|
myosin X-like 3 |
| chr2_+_30787128 | 0.05 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr5_+_24287927 | 0.05 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr1_+_41690402 | 0.05 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr2_-_48171441 | 0.05 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr1_-_45920632 | 0.05 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr23_-_21797517 | 0.05 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr7_-_49594995 | 0.04 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr19_+_24068223 | 0.04 |
ENSDART00000141351
ENSDART00000100420 |
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr12_+_17042754 | 0.04 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr24_-_38079261 | 0.04 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr13_+_12528043 | 0.04 |
ENSDART00000057761
|
rrh
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr18_+_49969568 | 0.03 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr2_-_48171112 | 0.03 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr16_+_50163352 | 0.03 |
ENSDART00000153890
ENSDART00000060522 |
si:ch211-231m23.4
|
si:ch211-231m23.4 |
| chr2_-_58075414 | 0.03 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr2_+_7192966 | 0.03 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr5_+_52067723 | 0.03 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr22_+_35275468 | 0.03 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr13_-_36525982 | 0.03 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr10_+_6013076 | 0.03 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr2_+_30531726 | 0.03 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr22_-_14696667 | 0.03 |
ENSDART00000180379
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr12_+_6195191 | 0.03 |
ENSDART00000043236
ENSDART00000186420 |
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr7_+_4384863 | 0.02 |
ENSDART00000042955
ENSDART00000134653 |
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr7_-_17780048 | 0.02 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr2_+_13050225 | 0.02 |
ENSDART00000182237
|
BX324233.1
|
|
| chr6_-_39653972 | 0.01 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr22_+_5103349 | 0.01 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr17_-_50010121 | 0.01 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr21_-_41369370 | 0.01 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr17_-_2690083 | 0.01 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr6_-_59381391 | 0.01 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr2_-_9696283 | 0.01 |
ENSDART00000165712
|
selenot1a
|
selenoprotein T, 1a |
| chr5_-_50748081 | 0.01 |
ENSDART00000113985
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr22_+_38164486 | 0.00 |
ENSDART00000137521
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr7_+_20031202 | 0.00 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr22_+_35205546 | 0.00 |
ENSDART00000189203
|
tsc22d2
|
TSC22 domain family 2 |
| chr18_+_19975787 | 0.00 |
ENSDART00000138103
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr7_-_12821277 | 0.00 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zgc:101100
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.6 | GO:0007602 | phototransduction(GO:0007602) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |