Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
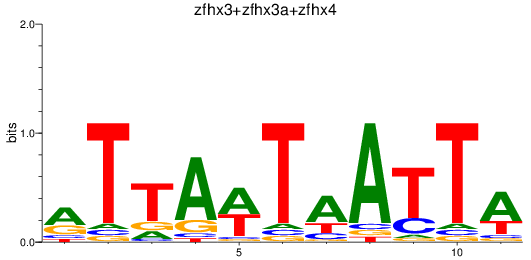
Results for zfhx3+zfhx3a+zfhx4
Z-value: 0.91
Transcription factors associated with zfhx3+zfhx3a+zfhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zfhx3a
|
ENSDARG00000073944 | si_ch73-386h18.1 |
|
zfhx4
|
ENSDARG00000075542 | zinc finger homeobox 4 |
|
zfhx3
|
ENSDARG00000103057 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch73-386h18.1 | dr11_v1_chr18_+_6054816_6054816 | -0.87 | 2.3e-03 | Click! |
| zfhx4 | dr11_v1_chr24_-_23323526_23323526 | -0.75 | 1.9e-02 | Click! |
| zfhx3 | dr11_v1_chr7_-_68373495_68373495 | -0.72 | 2.7e-02 | Click! |
Activity profile of zfhx3+zfhx3a+zfhx4 motif
Sorted Z-values of zfhx3+zfhx3a+zfhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_38299385 | 1.57 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr25_+_34915762 | 1.42 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr22_+_14836291 | 1.30 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr15_+_38299563 | 1.27 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr15_+_29472065 | 1.23 |
ENSDART00000154343
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr22_-_24791505 | 1.19 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr21_-_13123176 | 1.16 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr23_+_24589633 | 1.14 |
ENSDART00000143694
|
mlh3
|
mutL homolog 3 (E. coli) |
| chr16_-_27566552 | 1.13 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr8_-_20230559 | 1.13 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr22_+_15960514 | 1.11 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr9_+_45227028 | 1.08 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr25_+_28679672 | 1.06 |
ENSDART00000139965
ENSDART00000134072 |
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr9_-_14143370 | 1.00 |
ENSDART00000108950
|
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr8_+_10339869 | 0.98 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr19_-_3056235 | 0.97 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr12_+_46582727 | 0.95 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr13_+_25286538 | 0.95 |
ENSDART00000180282
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr22_+_15960005 | 0.93 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr5_-_34875858 | 0.89 |
ENSDART00000085086
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr22_+_15959844 | 0.87 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr5_+_19933356 | 0.86 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr19_+_2631565 | 0.85 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr8_-_20230802 | 0.85 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr12_+_27061720 | 0.82 |
ENSDART00000153426
|
srcap
|
Snf2-related CREBBP activator protein |
| chr12_+_30788912 | 0.81 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr18_+_7611298 | 0.81 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr22_-_24818066 | 0.79 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr6_-_43283122 | 0.78 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr11_+_24804556 | 0.77 |
ENSDART00000082761
ENSDART00000186283 |
adipor1a
|
adiponectin receptor 1a |
| chr5_-_19932621 | 0.77 |
ENSDART00000088881
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr18_+_16053455 | 0.77 |
ENSDART00000189163
ENSDART00000188269 |
FO834850.1
|
|
| chr8_-_25569920 | 0.75 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr5_+_9224051 | 0.74 |
ENSDART00000139265
|
si:ch211-12e13.12
|
si:ch211-12e13.12 |
| chr17_-_51893123 | 0.74 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr10_+_8197827 | 0.71 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr24_-_2450597 | 0.70 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr5_+_6954162 | 0.70 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr20_-_40754794 | 0.68 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr13_+_31205439 | 0.68 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr23_+_45200481 | 0.66 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr16_-_41787421 | 0.65 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr13_+_45524475 | 0.63 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr20_+_31076488 | 0.61 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr10_-_11261565 | 0.59 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr22_-_34551568 | 0.58 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr20_+_36629173 | 0.57 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr3_-_20040636 | 0.57 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr1_+_22851261 | 0.57 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr16_-_55259199 | 0.53 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr12_+_47081783 | 0.53 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr16_-_44127307 | 0.52 |
ENSDART00000104583
ENSDART00000151936 ENSDART00000058685 ENSDART00000190830 |
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr21_+_37513488 | 0.51 |
ENSDART00000185394
|
amot
|
angiomotin |
| chr12_-_18578218 | 0.51 |
ENSDART00000125803
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr25_+_34915576 | 0.50 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr16_-_31351419 | 0.50 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr20_+_30610547 | 0.50 |
ENSDART00000122256
|
ccr6a
|
chemokine (C-C motif) receptor 6a |
| chr2_+_29249561 | 0.49 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr15_-_3534705 | 0.48 |
ENSDART00000158150
|
cog6
|
component of oligomeric golgi complex 6 |
| chr12_-_18578432 | 0.46 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr10_-_8197049 | 0.45 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr5_+_13394543 | 0.45 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr17_+_8799451 | 0.45 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr19_+_3056450 | 0.44 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr21_-_39177564 | 0.44 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr20_+_27298783 | 0.42 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr10_-_11261386 | 0.40 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr6_-_48473694 | 0.40 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr17_+_8212477 | 0.39 |
ENSDART00000064665
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr24_+_26329018 | 0.39 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr22_+_2751887 | 0.39 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr3_+_29714775 | 0.39 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr5_+_28830388 | 0.38 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr8_+_50742975 | 0.38 |
ENSDART00000155664
ENSDART00000160612 |
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr17_-_30975978 | 0.38 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr4_-_22338906 | 0.37 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr23_+_13375359 | 0.37 |
ENSDART00000151947
|
ZNF512B
|
si:dkey-256k13.2 |
| chr8_+_1839695 | 0.36 |
ENSDART00000148254
ENSDART00000143473 |
snap29
|
synaptosomal-associated protein 29 |
| chr4_-_27117112 | 0.36 |
ENSDART00000034534
|
zbed4
|
zinc finger, BED-type containing 4 |
| chr5_-_30620625 | 0.35 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr24_+_26328787 | 0.35 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr16_+_24733741 | 0.34 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr18_-_17399291 | 0.32 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr2_-_30784198 | 0.32 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr5_+_28848870 | 0.32 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr5_+_28849155 | 0.31 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr17_-_24820801 | 0.31 |
ENSDART00000156606
|
trmt61b
|
tRNA methyltransferase 61B |
| chr1_+_26480890 | 0.30 |
ENSDART00000164430
|
uso1
|
USO1 vesicle transport factor |
| chr6_-_43498996 | 0.30 |
ENSDART00000154742
|
MDFIC2
|
si:dkey-58f6.2 |
| chr12_-_28848015 | 0.30 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr20_+_41756996 | 0.29 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr20_+_19423823 | 0.29 |
ENSDART00000152216
|
znf513a
|
zinc finger protein 513a |
| chr5_+_4054704 | 0.29 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr11_+_7432533 | 0.28 |
ENSDART00000180977
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr6_-_29515709 | 0.27 |
ENSDART00000180205
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr5_+_28858345 | 0.27 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr18_+_2228737 | 0.26 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr20_+_45620076 | 0.26 |
ENSDART00000113806
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr16_-_25608453 | 0.26 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr17_+_8799661 | 0.24 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr9_+_12444494 | 0.23 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr5_+_28830643 | 0.21 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr3_-_5228137 | 0.21 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr23_-_29394505 | 0.21 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr3_-_31715714 | 0.20 |
ENSDART00000051542
|
ccdc47
|
coiled-coil domain containing 47 |
| chr25_+_29474583 | 0.20 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr1_+_33558555 | 0.20 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr2_+_29249204 | 0.19 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr14_-_7207961 | 0.19 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr23_-_41965557 | 0.18 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr22_-_38480186 | 0.16 |
ENSDART00000171704
|
soul4
|
heme-binding protein soul4 |
| chr14_+_2487672 | 0.15 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr1_+_44003654 | 0.15 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr11_+_29770966 | 0.14 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr14_+_8594367 | 0.14 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| chr1_-_15797663 | 0.13 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr7_-_20865005 | 0.12 |
ENSDART00000190752
|
fis1
|
fission, mitochondrial 1 |
| chr2_-_7845110 | 0.12 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr1_+_44838706 | 0.12 |
ENSDART00000162779
ENSDART00000147357 |
kdm2aa
|
lysine (K)-specific demethylase 2Aa |
| chr18_+_15644559 | 0.10 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr7_-_17443332 | 0.08 |
ENSDART00000182348
|
nitr3c
|
novel immune-type receptor 3c |
| chr17_+_9310259 | 0.08 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr10_+_11261576 | 0.07 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr2_-_27609189 | 0.06 |
ENSDART00000040555
|
lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr18_-_22713621 | 0.06 |
ENSDART00000079013
|
si:ch73-113g13.2
|
si:ch73-113g13.2 |
| chr10_-_42131408 | 0.06 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr15_+_35139457 | 0.05 |
ENSDART00000059204
|
slc19a3a
|
solute carrier family 19 (thiamine transporter), member 3a |
| chr10_-_17179415 | 0.05 |
ENSDART00000131751
|
depdc5
|
DEP domain containing 5 |
| chr20_-_40750953 | 0.05 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr22_-_294700 | 0.05 |
ENSDART00000189179
|
CABZ01079179.1
|
|
| chr14_+_10596628 | 0.03 |
ENSDART00000115177
|
gpr174
|
G protein-coupled receptor 174 |
| chr14_-_25111496 | 0.03 |
ENSDART00000158108
|
si:rp71-1d10.8
|
si:rp71-1d10.8 |
| chr17_-_7371564 | 0.02 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr18_-_22713402 | 0.02 |
ENSDART00000145871
|
si:ch73-113g13.2
|
si:ch73-113g13.2 |
| chr17_+_2162916 | 0.02 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr23_+_20689255 | 0.01 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr18_+_29950233 | 0.01 |
ENSDART00000146431
|
atmin
|
ATM interactor |
| chr21_+_13150937 | 0.01 |
ENSDART00000102251
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr17_+_31914877 | 0.01 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr22_+_19220459 | 0.00 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zfhx3+zfhx3a+zfhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 1.0 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 1.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 1.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.3 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.3 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.5 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 2.0 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 0.4 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 1.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.1 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.5 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.1 | GO:0043653 | peroxisome fission(GO:0016559) mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.2 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0007530 | sex determination(GO:0007530) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 2.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.2 | 1.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 0.6 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.2 | 0.8 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.0 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.3 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |