Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for zbtb7b
Z-value: 0.48
Transcription factors associated with zbtb7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb7b
|
ENSDARG00000021891 | zinc finger and BTB domain containing 7B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb7b | dr11_v1_chr16_-_54810464_54810464 | 0.66 | 5.3e-02 | Click! |
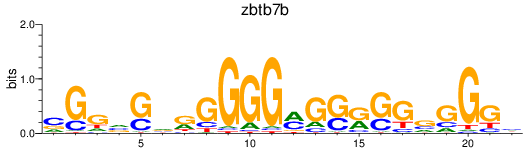
Activity profile of zbtb7b motif
Sorted Z-values of zbtb7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_26715655 | 0.82 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr4_-_77551860 | 0.68 |
ENSDART00000188176
|
AL935186.6
|
|
| chr9_-_8314028 | 0.64 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr18_-_41161828 | 0.61 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr4_-_77563411 | 0.59 |
ENSDART00000186841
|
AL935186.8
|
|
| chr14_-_24111292 | 0.58 |
ENSDART00000186611
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr17_-_8312923 | 0.56 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr8_-_52938439 | 0.55 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr10_-_25543227 | 0.53 |
ENSDART00000007778
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr13_+_45582391 | 0.46 |
ENSDART00000058093
|
ldlrap1b
|
low density lipoprotein receptor adaptor protein 1b |
| chr5_-_30080332 | 0.41 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr2_-_52550135 | 0.41 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr11_-_11878099 | 0.39 |
ENSDART00000188314
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr18_-_26715156 | 0.38 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr17_+_37310663 | 0.37 |
ENSDART00000157122
|
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr1_-_59216197 | 0.36 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr17_+_26965351 | 0.36 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr20_+_39250673 | 0.36 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr1_+_14454663 | 0.35 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr19_-_2707048 | 0.35 |
ENSDART00000166112
|
il6
|
interleukin 6 (interferon, beta 2) |
| chr10_+_44057177 | 0.34 |
ENSDART00000164610
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr17_+_12075805 | 0.34 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr10_+_44057502 | 0.34 |
ENSDART00000183868
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr1_+_15062 | 0.34 |
ENSDART00000169005
|
cep97
|
centrosomal protein 97 |
| chr6_-_29377092 | 0.33 |
ENSDART00000078665
|
tmem131
|
transmembrane protein 131 |
| chr1_+_44196236 | 0.33 |
ENSDART00000179560
ENSDART00000166324 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr1_+_29741843 | 0.33 |
ENSDART00000136066
|
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
| chr2_+_29884363 | 0.32 |
ENSDART00000139247
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr23_+_39346774 | 0.31 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr22_-_20403194 | 0.31 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr21_-_11646878 | 0.31 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr23_+_39346930 | 0.30 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr7_+_19552381 | 0.27 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr19_+_13994563 | 0.27 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr6_-_53326421 | 0.27 |
ENSDART00000191740
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr6_+_3334392 | 0.27 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr15_+_44184367 | 0.27 |
ENSDART00000162918
ENSDART00000110060 |
zgc:165514
|
zgc:165514 |
| chr20_-_9194257 | 0.26 |
ENSDART00000133012
|
ylpm1
|
YLP motif containing 1 |
| chr3_-_29891456 | 0.26 |
ENSDART00000151677
ENSDART00000014021 |
slc25a39
|
solute carrier family 25, member 39 |
| chr21_-_18932761 | 0.26 |
ENSDART00000140129
|
med15
|
mediator complex subunit 15 |
| chr22_-_7050 | 0.26 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr5_-_30074332 | 0.26 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr14_+_31498439 | 0.25 |
ENSDART00000173281
|
phf6
|
PHD finger protein 6 |
| chr3_+_3810919 | 0.25 |
ENSDART00000056035
|
FQ311927.1
|
|
| chr1_-_8012476 | 0.25 |
ENSDART00000177051
|
CR855320.3
|
|
| chr8_+_8947623 | 0.25 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr22_+_26703026 | 0.24 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr1_+_7414318 | 0.24 |
ENSDART00000127426
|
si:ch73-383l1.1
|
si:ch73-383l1.1 |
| chr17_+_23730089 | 0.24 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr6_-_10791880 | 0.24 |
ENSDART00000104884
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr10_+_1849874 | 0.23 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr15_-_47607480 | 0.23 |
ENSDART00000171605
|
socs9
|
suppressor of cytokine signaling 9 |
| chr16_-_34212912 | 0.22 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr14_-_16423200 | 0.22 |
ENSDART00000108868
ENSDART00000161793 |
maml1
|
mastermind-like transcriptional coactivator 1 |
| chr12_+_10443785 | 0.21 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr15_-_25198107 | 0.21 |
ENSDART00000159793
|
mnta
|
MAX network transcriptional repressor a |
| chr7_-_5070794 | 0.21 |
ENSDART00000097877
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr20_-_54435287 | 0.20 |
ENSDART00000148632
|
yy1b
|
YY1 transcription factor b |
| chr21_-_45871866 | 0.20 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr21_-_2223771 | 0.20 |
ENSDART00000158880
ENSDART00000162725 |
zgc:113343
|
zgc:113343 |
| chr6_+_28203 | 0.20 |
ENSDART00000191561
|
CZQB01141835.1
|
|
| chr12_+_40945319 | 0.19 |
ENSDART00000183435
|
CDH18
|
cadherin 18 |
| chr8_-_5847533 | 0.19 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr5_+_37744625 | 0.18 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr10_+_22034477 | 0.18 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr16_-_8120203 | 0.18 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr12_+_48841419 | 0.18 |
ENSDART00000125331
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr4_-_74109792 | 0.18 |
ENSDART00000174320
|
RAB21
|
zgc:154045 |
| chr17_+_43946269 | 0.18 |
ENSDART00000017376
|
ktn1
|
kinectin 1 |
| chr4_-_2380173 | 0.18 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr1_+_594584 | 0.18 |
ENSDART00000135944
|
jam2a
|
junctional adhesion molecule 2a |
| chr20_+_35058634 | 0.18 |
ENSDART00000122696
|
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr21_-_233282 | 0.17 |
ENSDART00000157684
|
bxdc2
|
brix domain containing 2 |
| chr8_-_53488832 | 0.17 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr17_+_23729717 | 0.17 |
ENSDART00000179026
|
zgc:91976
|
zgc:91976 |
| chr12_-_14211293 | 0.17 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr17_-_8976307 | 0.17 |
ENSDART00000092113
|
zranb1b
|
zinc finger, RAN-binding domain containing 1b |
| chr5_-_35264517 | 0.17 |
ENSDART00000114981
|
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr3_-_23643751 | 0.17 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr18_-_2727764 | 0.17 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr2_-_44746723 | 0.17 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium chain family member 3 |
| chr15_+_42397125 | 0.17 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr14_-_7207961 | 0.16 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr25_+_4581214 | 0.16 |
ENSDART00000185552
|
CABZ01068600.1
|
|
| chr8_+_35356944 | 0.16 |
ENSDART00000143243
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr20_+_474288 | 0.16 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr1_-_20970266 | 0.15 |
ENSDART00000088545
|
klhl2
|
kelch-like family member 2 |
| chr24_-_11467004 | 0.15 |
ENSDART00000144316
|
pxdc1b
|
PX domain containing 1b |
| chr6_-_4214297 | 0.15 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr11_-_23322182 | 0.15 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr4_+_357810 | 0.15 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr14_-_48245816 | 0.14 |
ENSDART00000172452
|
rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr24_-_36593876 | 0.14 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr6_-_43677125 | 0.14 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr6_+_52918537 | 0.14 |
ENSDART00000174229
|
or137-1
|
odorant receptor, family H, subfamily 137, member 1 |
| chr17_-_4395373 | 0.14 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr17_-_2721336 | 0.14 |
ENSDART00000109582
ENSDART00000192691 ENSDART00000189381 |
spata7
|
spermatogenesis associated 7 |
| chr11_-_9948487 | 0.13 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr13_-_50366950 | 0.13 |
ENSDART00000098209
|
sirt1
|
sirtuin 1 |
| chr13_-_4367083 | 0.13 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr3_+_22079219 | 0.13 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr18_-_8030073 | 0.13 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr22_-_506522 | 0.13 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr24_+_37825634 | 0.12 |
ENSDART00000129889
|
ift140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr24_+_42131564 | 0.12 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr10_-_5857548 | 0.12 |
ENSDART00000166933
|
si:ch211-281k19.2
|
si:ch211-281k19.2 |
| chr9_-_704667 | 0.12 |
ENSDART00000147092
|
cflarb
|
CASP8 and FADD-like apoptosis regulator b |
| chr11_-_40504170 | 0.12 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr20_-_21672970 | 0.12 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr20_+_27242595 | 0.12 |
ENSDART00000149300
|
si:dkey-85n7.6
|
si:dkey-85n7.6 |
| chr13_-_24218795 | 0.12 |
ENSDART00000136217
|
galnt2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 |
| chr11_-_19501439 | 0.12 |
ENSDART00000158328
ENSDART00000103983 |
slc2a9l1
|
solute carrier family 2 (facilitated glucose transporter), member 9-like 1 |
| chr15_-_5742531 | 0.12 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr3_+_2669813 | 0.11 |
ENSDART00000014205
|
CR388047.1
|
|
| chr3_-_56933578 | 0.11 |
ENSDART00000192185
|
hid1a
|
HID1 domain containing a |
| chr21_+_1143141 | 0.11 |
ENSDART00000178294
|
CABZ01086139.1
|
|
| chr5_-_69621227 | 0.11 |
ENSDART00000178543
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr2_+_24781026 | 0.11 |
ENSDART00000145692
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr16_+_7991274 | 0.11 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr16_-_17586883 | 0.11 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr21_+_233271 | 0.11 |
ENSDART00000171440
|
dtwd2
|
DTW domain containing 2 |
| chr16_+_23531583 | 0.11 |
ENSDART00000146708
|
adar
|
adenosine deaminase, RNA-specific |
| chr9_+_29323163 | 0.10 |
ENSDART00000184908
|
oxgr1b
|
oxoglutarate (alpha-ketoglutarate) receptor 1b |
| chr15_-_37543839 | 0.10 |
ENSDART00000148870
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr24_+_15020402 | 0.10 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr25_-_35143360 | 0.10 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr12_-_4475890 | 0.10 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr15_-_47956388 | 0.09 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr4_+_65306260 | 0.09 |
ENSDART00000167558
|
si:dkey-14o6.8
|
si:dkey-14o6.8 |
| chr25_-_19395156 | 0.09 |
ENSDART00000155335
|
map1ab
|
microtubule-associated protein 1Ab |
| chr20_-_2355357 | 0.09 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr22_-_6219674 | 0.09 |
ENSDART00000182582
|
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr21_+_15601100 | 0.08 |
ENSDART00000180558
ENSDART00000145454 |
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr2_+_31475772 | 0.08 |
ENSDART00000130722
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr23_+_22335407 | 0.08 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr7_-_56835543 | 0.08 |
ENSDART00000010322
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr5_-_31928913 | 0.08 |
ENSDART00000142919
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr19_-_6982272 | 0.08 |
ENSDART00000141913
ENSDART00000150957 |
znf384l
|
zinc finger protein 384 like |
| chr11_-_31146808 | 0.08 |
ENSDART00000172781
|
si:cabz01069582.1
|
si:cabz01069582.1 |
| chr19_-_11237125 | 0.08 |
ENSDART00000163921
|
ssr2
|
signal sequence receptor, beta |
| chr3_-_16289826 | 0.08 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr6_-_57426097 | 0.08 |
ENSDART00000171531
|
znfx1
|
zinc finger, NFX1-type containing 1 |
| chr18_+_32780450 | 0.08 |
ENSDART00000168399
|
olfcg7
|
olfactory receptor C family, g7 |
| chr6_+_52247993 | 0.08 |
ENSDART00000172176
|
zc3h10
|
zinc finger CCCH-type containing 10 |
| chr22_-_17729778 | 0.07 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr5_-_41142467 | 0.07 |
ENSDART00000129415
|
zfr
|
zinc finger RNA binding protein |
| chr9_-_32191620 | 0.07 |
ENSDART00000139426
|
pimr143
|
Pim proto-oncogene, serine/threonine kinase, related 143 |
| chr19_-_5058908 | 0.07 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr5_-_13645995 | 0.07 |
ENSDART00000099665
ENSDART00000166957 |
purba
|
purine-rich element binding protein Ba |
| chr5_-_41142768 | 0.07 |
ENSDART00000074789
|
zfr
|
zinc finger RNA binding protein |
| chr23_+_19182819 | 0.07 |
ENSDART00000131804
|
si:dkey-93l1.4
|
si:dkey-93l1.4 |
| chr1_+_594736 | 0.07 |
ENSDART00000166731
|
jam2a
|
junctional adhesion molecule 2a |
| chr25_-_19395476 | 0.07 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr2_+_258698 | 0.07 |
ENSDART00000181330
ENSDART00000181645 |
phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr22_+_413349 | 0.07 |
ENSDART00000082453
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr6_-_425378 | 0.07 |
ENSDART00000192190
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr2_+_24374305 | 0.07 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr23_-_44848961 | 0.06 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr16_-_12470034 | 0.06 |
ENSDART00000147483
|
ephb6
|
eph receptor B6 |
| chr1_+_41498188 | 0.06 |
ENSDART00000191934
ENSDART00000146310 |
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr19_+_935565 | 0.06 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr8_-_22838364 | 0.06 |
ENSDART00000187954
ENSDART00000185368 ENSDART00000188210 |
magixa
|
MAGI family member, X-linked a |
| chr14_-_15154695 | 0.06 |
ENSDART00000160677
|
uvssa
|
UV-stimulated scaffold protein A |
| chr9_-_37637179 | 0.06 |
ENSDART00000188143
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr5_-_10012971 | 0.06 |
ENSDART00000179421
|
CR936408.2
|
|
| chr21_+_19925910 | 0.06 |
ENSDART00000111694
ENSDART00000132653 |
tnksa
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase a |
| chr18_+_33570170 | 0.06 |
ENSDART00000133276
|
si:dkey-47k20.3
|
si:dkey-47k20.3 |
| chr9_-_264173 | 0.05 |
ENSDART00000166231
ENSDART00000165585 |
pcbp2
|
poly(rC) binding protein 2 |
| chr24_+_27023616 | 0.05 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr13_-_27767330 | 0.05 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr4_-_22472653 | 0.05 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr3_-_4501026 | 0.05 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr4_-_25812329 | 0.05 |
ENSDART00000146658
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr21_+_13577227 | 0.05 |
ENSDART00000146283
|
si:dkey-248f6.3
|
si:dkey-248f6.3 |
| chr22_+_12366516 | 0.05 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr15_+_3125136 | 0.05 |
ENSDART00000130968
|
rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr25_-_34284144 | 0.05 |
ENSDART00000188109
|
gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr13_-_51922290 | 0.04 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr24_-_38110779 | 0.04 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr18_+_30370339 | 0.04 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr16_+_16826504 | 0.04 |
ENSDART00000108691
|
kcnj14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr18_+_30370559 | 0.04 |
ENSDART00000165450
|
gse1
|
Gse1 coiled-coil protein |
| chr23_+_19691146 | 0.04 |
ENSDART00000143001
|
si:dkey-93l1.6
|
si:dkey-93l1.6 |
| chr19_+_18903533 | 0.04 |
ENSDART00000157523
ENSDART00000166562 |
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr10_-_20481763 | 0.04 |
ENSDART00000091156
ENSDART00000192358 |
tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr2_+_24374669 | 0.04 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr7_-_5711197 | 0.04 |
ENSDART00000134367
|
si:dkey-10h3.2
|
si:dkey-10h3.2 |
| chr5_-_26936861 | 0.04 |
ENSDART00000191032
|
htra4
|
HtrA serine peptidase 4 |
| chr25_+_4750972 | 0.04 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr7_-_38477235 | 0.04 |
ENSDART00000084355
|
zgc:165481
|
zgc:165481 |
| chr2_-_23479714 | 0.04 |
ENSDART00000167291
|
si:ch211-14p21.3
|
si:ch211-14p21.3 |
| chr1_+_56779495 | 0.04 |
ENSDART00000192871
|
FO680692.1
|
|
| chr6_+_3334710 | 0.04 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr4_-_75505048 | 0.04 |
ENSDART00000163665
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr9_+_51891 | 0.04 |
ENSDART00000163529
|
zgc:158316
|
zgc:158316 |
| chr22_-_367569 | 0.03 |
ENSDART00000041895
|
ssu72
|
SSU72 homolog, RNA polymerase II CTD phosphatase |
| chr7_+_34453185 | 0.03 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr22_-_24900121 | 0.03 |
ENSDART00000147643
|
si:dkey-4c23.3
|
si:dkey-4c23.3 |
| chr3_+_52467879 | 0.03 |
ENSDART00000156039
|
adgre5a
|
adhesion G protein-coupled receptor E5a |
| chr23_-_14057191 | 0.03 |
ENSDART00000154908
|
adcy6a
|
adenylate cyclase 6a |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.6 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.7 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.4 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 0.7 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 0.2 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.6 | GO:0003128 | heart field specification(GO:0003128) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.2 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.0 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0007520 | myoblast fusion(GO:0007520) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.7 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.1 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |