Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
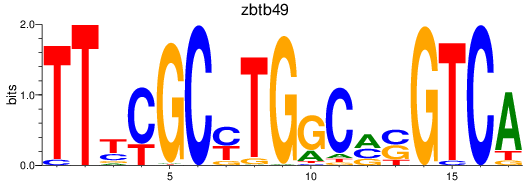
Results for zbtb49
Z-value: 0.48
Transcription factors associated with zbtb49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb49
|
ENSDARG00000102111 | zinc finger and BTB domain containing 49 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb49 | dr11_v1_chr14_-_30050_30050 | -0.68 | 4.6e-02 | Click! |
Activity profile of zbtb49 motif
Sorted Z-values of zbtb49 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_10120442 | 1.46 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr18_-_44129151 | 1.16 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| chr24_-_3419998 | 1.03 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr1_-_38709551 | 1.01 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr11_+_8565323 | 0.96 |
ENSDART00000081607
ENSDART00000141612 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr7_+_22823889 | 0.90 |
ENSDART00000127467
ENSDART00000148576 ENSDART00000149993 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr11_+_8565828 | 0.86 |
ENSDART00000126523
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr8_-_38477817 | 0.60 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr8_+_25302172 | 0.50 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr14_+_33882973 | 0.49 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr9_+_8942258 | 0.49 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr7_-_38340674 | 0.46 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr7_-_5148388 | 0.44 |
ENSDART00000143549
|
si:ch211-272h9.5
|
si:ch211-272h9.5 |
| chr17_+_8925232 | 0.43 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr9_+_14152211 | 0.40 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr9_+_38629962 | 0.32 |
ENSDART00000184890
|
znf148
|
zinc finger protein 148 |
| chr10_-_15919839 | 0.28 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr13_+_51853716 | 0.20 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr7_-_31941670 | 0.20 |
ENSDART00000180929
ENSDART00000075389 |
bdnf
|
brain-derived neurotrophic factor |
| chr21_+_25181003 | 0.19 |
ENSDART00000169700
|
si:dkey-183i3.9
|
si:dkey-183i3.9 |
| chr7_-_44794322 | 0.13 |
ENSDART00000019270
|
nae1
|
nedd8 activating enzyme E1 subunit 1 |
| chr6_+_36878815 | 0.13 |
ENSDART00000192482
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr22_+_12798569 | 0.13 |
ENSDART00000005720
|
stat1a
|
signal transducer and activator of transcription 1a |
| chr14_-_17622080 | 0.11 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr7_-_52334840 | 0.11 |
ENSDART00000174173
|
CR938716.1
|
|
| chr21_-_22673758 | 0.08 |
ENSDART00000164910
|
gig2i
|
grass carp reovirus (GCRV)-induced gene 2i |
| chr22_-_8455462 | 0.04 |
ENSDART00000185238
|
CABZ01061508.1
|
|
| chr14_+_9581896 | 0.02 |
ENSDART00000114563
|
tmem129
|
transmembrane protein 129, E3 ubiquitin protein ligase |
| chr7_-_31941330 | 0.01 |
ENSDART00000144682
|
bdnf
|
brain-derived neurotrophic factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb49
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 0.5 | GO:0042940 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.1 | 1.5 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 1.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.6 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 1.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.6 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |