Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
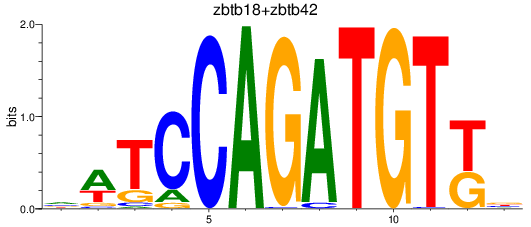
Results for zbtb18+zbtb42
Z-value: 1.70
Transcription factors associated with zbtb18+zbtb42
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb18
|
ENSDARG00000028228 | zinc finger and BTB domain containing 18 |
|
zbtb42
|
ENSDARG00000102761 | zinc finger and BTB domain containing 42 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb18 | dr11_v1_chr13_+_11436130_11436130 | 0.62 | 7.3e-02 | Click! |
| zbtb42 | dr11_v1_chr17_-_1407593_1407725 | -0.38 | 3.1e-01 | Click! |
Activity profile of zbtb18+zbtb42 motif
Sorted Z-values of zbtb18+zbtb42 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_26282672 | 3.58 |
ENSDART00000176663
|
and1
|
actinodin1 |
| chr25_+_31277415 | 3.01 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr23_+_21473103 | 2.78 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr23_-_21453614 | 2.69 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr1_+_31864404 | 2.59 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr25_+_31276842 | 2.56 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr11_-_24681292 | 2.53 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr1_+_45351890 | 2.45 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr23_-_21471022 | 2.36 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr10_-_29903165 | 2.35 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr23_+_21459263 | 2.31 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr20_+_23238833 | 2.30 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr25_+_35502552 | 2.28 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr21_-_19006631 | 2.20 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr4_-_16345227 | 2.16 |
ENSDART00000079521
|
kera
|
keratocan |
| chr23_-_21463788 | 2.07 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr15_+_33989181 | 2.03 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr10_-_22803740 | 1.90 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr13_+_12045475 | 1.68 |
ENSDART00000163053
ENSDART00000160812 ENSDART00000158244 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr18_+_402048 | 1.64 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr3_-_53486169 | 1.57 |
ENSDART00000115243
|
soul5
|
heme-binding protein soul5 |
| chr18_+_48423973 | 1.57 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr8_-_17064243 | 1.57 |
ENSDART00000185313
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr20_+_16750177 | 1.56 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr19_+_32166702 | 1.53 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr7_+_34290051 | 1.53 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr21_-_20341836 | 1.46 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr23_-_27857051 | 1.44 |
ENSDART00000043462
ENSDART00000137861 |
acvrl1
|
activin A receptor like type 1 |
| chr9_-_98982 | 1.44 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr10_+_21737745 | 1.41 |
ENSDART00000170498
ENSDART00000167997 |
pcdh1g18
|
protocadherin 1 gamma 18 |
| chr16_-_13595027 | 1.38 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr16_+_42830152 | 1.38 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr20_-_40451115 | 1.37 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr19_+_47311020 | 1.37 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr20_-_47704973 | 1.36 |
ENSDART00000174808
|
tfap2b
|
transcription factor AP-2 beta |
| chr25_-_15040369 | 1.33 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr10_-_24371312 | 1.33 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr6_+_22597362 | 1.32 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr1_-_21409877 | 1.31 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr25_-_34632050 | 1.31 |
ENSDART00000170671
|
megf8
|
multiple EGF-like-domains 8 |
| chr15_-_34567370 | 1.30 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr24_-_37877743 | 1.28 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr19_+_22216778 | 1.27 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr21_-_20342096 | 1.22 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr19_-_44091405 | 1.22 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr16_+_42829735 | 1.21 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr19_-_5699703 | 1.20 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr17_+_42274825 | 1.19 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr3_-_36839115 | 1.17 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr2_+_26647472 | 1.16 |
ENSDART00000145415
ENSDART00000157409 |
ttpa
|
tocopherol (alpha) transfer protein |
| chr13_-_31452516 | 1.15 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr20_+_2460864 | 1.14 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr16_-_26676685 | 1.11 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr4_+_4232562 | 1.07 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr24_-_28243186 | 1.06 |
ENSDART00000105691
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr7_+_29133321 | 1.04 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr9_-_38587275 | 1.03 |
ENSDART00000077446
|
si:dkey-101k6.5
|
si:dkey-101k6.5 |
| chr12_-_28881638 | 0.97 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr9_-_9842149 | 0.97 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr1_-_11973341 | 0.94 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr9_-_12424231 | 0.93 |
ENSDART00000188952
|
zgc:162707
|
zgc:162707 |
| chr6_+_1787160 | 0.91 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr9_-_12424791 | 0.91 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr18_-_6982499 | 0.91 |
ENSDART00000101525
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr10_+_8968203 | 0.89 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr7_+_17229980 | 0.87 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr24_-_982443 | 0.86 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr15_-_14552101 | 0.86 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr2_+_36646451 | 0.86 |
ENSDART00000039174
|
klhl6
|
kelch-like family member 6 |
| chr4_+_12612145 | 0.84 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr19_+_5315987 | 0.83 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr5_-_39474235 | 0.81 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr16_+_5579744 | 0.81 |
ENSDART00000147973
|
macf1b
|
microtubule-actin crosslinking factor 1 b |
| chr17_+_42274240 | 0.80 |
ENSDART00000134377
|
pax1a
|
paired box 1a |
| chr18_-_46241775 | 0.80 |
ENSDART00000145999
ENSDART00000134244 ENSDART00000185021 ENSDART00000181855 |
prx
|
periaxin |
| chr23_-_9919959 | 0.79 |
ENSDART00000127029
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr6_-_39270851 | 0.78 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr16_+_9609721 | 0.77 |
ENSDART00000047920
|
enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr12_+_24344963 | 0.76 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr14_+_21106444 | 0.76 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr18_+_17660158 | 0.75 |
ENSDART00000186279
|
cpne2
|
copine II |
| chr10_-_15128771 | 0.74 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr21_-_5879897 | 0.74 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr21_-_33995213 | 0.73 |
ENSDART00000140184
|
EBF1 (1 of many)
|
si:ch211-51e8.2 |
| chr16_+_8716800 | 0.72 |
ENSDART00000124693
ENSDART00000181961 |
cabz01093075.1
|
cabz01093075.1 |
| chr19_-_22478888 | 0.70 |
ENSDART00000090679
|
pleca
|
plectin a |
| chr19_+_19489588 | 0.70 |
ENSDART00000166755
|
tril
|
TLR4 interactor with leucine-rich repeats |
| chr20_+_6590220 | 0.70 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr4_+_9609905 | 0.69 |
ENSDART00000142284
ENSDART00000150687 |
meig1
|
meiosis/spermiogenesis associated 1 |
| chr9_-_44983666 | 0.69 |
ENSDART00000149704
|
itgb2
|
integrin, beta 2 |
| chr19_-_24267823 | 0.68 |
ENSDART00000132430
|
s100v2
|
S100 calcium binding protein V2 |
| chr13_-_13294847 | 0.67 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr19_+_24882845 | 0.67 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr24_-_28245872 | 0.66 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr10_+_7102461 | 0.66 |
ENSDART00000187539
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr4_+_12612723 | 0.66 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr17_+_29345606 | 0.65 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr7_-_2090594 | 0.62 |
ENSDART00000183166
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr13_+_18545819 | 0.62 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr5_-_31904562 | 0.61 |
ENSDART00000140640
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr5_-_22082918 | 0.61 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr19_-_35596207 | 0.61 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr11_+_10548171 | 0.60 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr15_-_30816370 | 0.60 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr16_+_54875530 | 0.60 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr23_+_19977120 | 0.60 |
ENSDART00000089342
|
cfap126
|
cilia and flagella associated protein 126 |
| chr24_-_37877554 | 0.59 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr7_-_25895189 | 0.58 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr21_+_18313152 | 0.58 |
ENSDART00000170510
|
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr6_-_13408680 | 0.58 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr8_+_38417461 | 0.57 |
ENSDART00000132718
|
nkx6.3
|
NK6 homeobox 3 |
| chr10_-_11761927 | 0.57 |
ENSDART00000138748
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr3_+_17537352 | 0.56 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr7_+_17229282 | 0.56 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr5_-_23429228 | 0.53 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr13_+_25422009 | 0.53 |
ENSDART00000057686
|
calhm2
|
calcium homeostasis modulator 2 |
| chr3_-_32596394 | 0.53 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr12_-_34716037 | 0.52 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr12_+_13652747 | 0.52 |
ENSDART00000066359
|
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr18_+_17660402 | 0.51 |
ENSDART00000143475
|
cpne2
|
copine II |
| chr20_+_50852356 | 0.50 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr15_-_8517376 | 0.50 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr9_+_33009284 | 0.50 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr8_-_25247284 | 0.48 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr6_+_51824596 | 0.47 |
ENSDART00000149003
|
rspo4
|
R-spondin 4 |
| chr17_+_38255105 | 0.46 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr6_+_612330 | 0.46 |
ENSDART00000166872
ENSDART00000191758 |
kynu
|
kynureninase |
| chr8_+_7033049 | 0.46 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr7_-_38612230 | 0.46 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr3_-_39363065 | 0.45 |
ENSDART00000155894
|
arhgap23a
|
Rho GTPase activating protein 23a |
| chr12_-_7806007 | 0.44 |
ENSDART00000190359
|
ank3b
|
ankyrin 3b |
| chr5_-_52752514 | 0.43 |
ENSDART00000190162
|
fam189a2
|
family with sequence similarity 189, member A2 |
| chr20_+_47434709 | 0.41 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr10_+_40836378 | 0.41 |
ENSDART00000085792
|
trim69
|
tripartite motif containing 69 |
| chr20_-_20932760 | 0.41 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr3_-_3731563 | 0.40 |
ENSDART00000180978
|
BX908804.2
|
|
| chr23_+_26079467 | 0.40 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr2_+_5793908 | 0.40 |
ENSDART00000145219
|
slc35d1b
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1b |
| chr16_+_21738194 | 0.40 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr25_+_15841670 | 0.40 |
ENSDART00000049992
|
syt9b
|
synaptotagmin IXb |
| chr3_-_54669185 | 0.40 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr2_-_51634431 | 0.39 |
ENSDART00000165568
|
pigr
|
polymeric immunoglobulin receptor |
| chr16_-_17300030 | 0.39 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr10_+_28587446 | 0.39 |
ENSDART00000030138
ENSDART00000137508 |
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr20_-_13673566 | 0.39 |
ENSDART00000181641
|
TAGAP
|
si:ch211-122h15.4 |
| chr2_-_37043540 | 0.38 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr4_-_71110826 | 0.37 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr2_+_11685742 | 0.36 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr2_+_31671545 | 0.36 |
ENSDART00000145446
|
ackr4a
|
atypical chemokine receptor 4a |
| chr10_+_20070178 | 0.35 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr22_-_5933844 | 0.34 |
ENSDART00000163370
ENSDART00000189331 |
si:rp71-36a1.2
|
si:rp71-36a1.2 |
| chr13_+_25421531 | 0.34 |
ENSDART00000158093
|
calhm2
|
calcium homeostasis modulator 2 |
| chr6_-_50704689 | 0.33 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr1_+_157793 | 0.32 |
ENSDART00000152205
|
cul4a
|
cullin 4A |
| chr24_+_41610068 | 0.32 |
ENSDART00000159507
|
CABZ01044108.1
|
|
| chr3_+_44947355 | 0.32 |
ENSDART00000083040
|
hs3st2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr4_-_9609634 | 0.31 |
ENSDART00000067188
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr19_+_27339848 | 0.30 |
ENSDART00000052355
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr4_-_1015896 | 0.30 |
ENSDART00000170292
|
FAM180A
|
family with sequence similarity 180 member A |
| chr5_+_19314574 | 0.30 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr19_+_47299212 | 0.30 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr4_-_49827366 | 0.29 |
ENSDART00000183761
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr20_-_8443425 | 0.29 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr7_-_71837213 | 0.29 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr6_-_36182115 | 0.27 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr12_+_20699534 | 0.27 |
ENSDART00000131883
|
si:ch211-119c20.2
|
si:ch211-119c20.2 |
| chr9_+_7456076 | 0.26 |
ENSDART00000125824
ENSDART00000122526 |
tmem198a
|
transmembrane protein 198a |
| chr24_-_1303553 | 0.26 |
ENSDART00000190984
|
nrp1a
|
neuropilin 1a |
| chr6_-_10320676 | 0.26 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr2_-_37043905 | 0.26 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr7_+_5494565 | 0.26 |
ENSDART00000135271
|
MYADM
|
si:dkeyp-67a8.2 |
| chr3_+_17846890 | 0.26 |
ENSDART00000193384
|
znf385c
|
zinc finger protein 385C |
| chr21_-_20381481 | 0.26 |
ENSDART00000115236
|
atp5mea
|
ATP synthase membrane subunit ea |
| chr5_-_19861766 | 0.25 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr7_-_56606752 | 0.23 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr5_+_19309877 | 0.23 |
ENSDART00000190338
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr8_-_29706882 | 0.23 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr17_-_30839338 | 0.23 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr11_-_37411492 | 0.22 |
ENSDART00000166468
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr3_+_33367954 | 0.22 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr18_-_21097663 | 0.22 |
ENSDART00000060196
|
anpepa
|
alanyl (membrane) aminopeptidase a |
| chr5_+_30741730 | 0.22 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr25_-_19661198 | 0.21 |
ENSDART00000149641
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr22_-_6884981 | 0.21 |
ENSDART00000124219
|
FO904898.4
|
|
| chr9_+_22764235 | 0.20 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr3_+_15296824 | 0.19 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr13_-_33170733 | 0.19 |
ENSDART00000057382
|
fbln5
|
fibulin 5 |
| chr3_+_19732173 | 0.18 |
ENSDART00000178684
|
mrc2
|
mannose receptor, C type 2 |
| chr9_+_29603649 | 0.17 |
ENSDART00000140477
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr7_-_41403022 | 0.17 |
ENSDART00000174285
|
BX322787.1
|
|
| chr20_-_38446891 | 0.16 |
ENSDART00000192013
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr16_+_2843428 | 0.16 |
ENSDART00000149485
|
clec3ba
|
C-type lectin domain family 3, member Ba |
| chr23_-_452365 | 0.16 |
ENSDART00000146776
|
tspan2b
|
tetraspanin 2b |
| chr14_+_48964628 | 0.16 |
ENSDART00000105427
|
mif
|
macrophage migration inhibitory factor |
| chr20_-_9278252 | 0.15 |
ENSDART00000136079
|
syt14b
|
synaptotagmin XIVb |
| chr24_+_34113424 | 0.15 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr22_-_8692305 | 0.15 |
ENSDART00000181602
|
CR450686.4
|
|
| chr24_-_1303935 | 0.15 |
ENSDART00000159212
ENSDART00000159267 ENSDART00000164904 |
nrp1a
|
neuropilin 1a |
| chr13_-_25198025 | 0.14 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr20_-_14875308 | 0.14 |
ENSDART00000141290
|
dnm3a
|
dynamin 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb18+zbtb42
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.4 | 1.4 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.2 | 0.9 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.2 | 1.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 1.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 1.4 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.2 | 0.5 | GO:0072579 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.6 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 1.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.4 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.5 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.8 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 5.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.1 | 1.3 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 2.1 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 2.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.6 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 1.2 | GO:0006775 | fat-soluble vitamin metabolic process(GO:0006775) |
| 0.1 | 0.8 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.0 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 2.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 1.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.5 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 1.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 2.3 | GO:0033333 | fin development(GO:0033333) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) neuron fate specification(GO:0048665) |
| 0.0 | 0.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 1.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 2.4 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.2 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 2.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 1.4 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 1.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 2.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 5.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 21.4 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.4 | 2.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.3 | 1.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 1.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.0 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.6 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 1.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 0.5 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 1.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.9 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.4 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 1.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 2.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.4 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.3 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.8 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 1.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 2.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 1.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 8.4 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.2 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |