Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
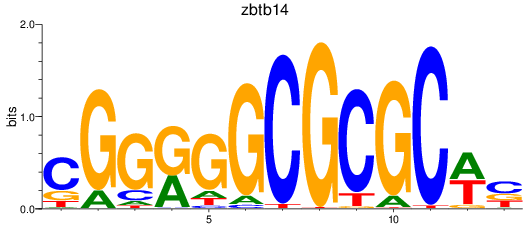
Results for zbtb14
Z-value: 2.41
Transcription factors associated with zbtb14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb14
|
ENSDARG00000098273 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb14 | dr11_v1_chr24_+_41989108_41989108 | -0.71 | 3.2e-02 | Click! |
Activity profile of zbtb14 motif
Sorted Z-values of zbtb14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22281854 | 9.01 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr14_+_46313135 | 6.19 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr24_+_32176155 | 4.78 |
ENSDART00000003745
|
vim
|
vimentin |
| chr19_+_25649626 | 4.08 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr2_+_42724404 | 3.22 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr14_+_46313396 | 3.08 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr4_+_5741733 | 2.99 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr1_-_56223913 | 2.86 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr23_-_20258490 | 2.80 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr23_+_44732863 | 2.72 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr14_+_15064870 | 2.58 |
ENSDART00000167075
|
nkx1.2lb
|
NK1 transcription factor related 2-like,b |
| chr20_-_40451115 | 2.42 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr19_-_5254699 | 2.39 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr25_-_22639133 | 2.35 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr21_+_11468934 | 2.29 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr19_+_3653976 | 2.26 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr14_-_25577094 | 2.22 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr13_+_1872767 | 2.21 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr25_+_35502552 | 2.21 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr4_+_22680442 | 2.19 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr22_-_13042992 | 2.14 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr16_-_43026273 | 2.11 |
ENSDART00000156820
ENSDART00000189080 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr1_+_53945934 | 2.11 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr16_-_43025885 | 2.11 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr22_-_372723 | 2.06 |
ENSDART00000112895
|
FNDC10
|
si:zfos-1324h11.5 |
| chr2_+_26179096 | 2.03 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr14_-_2163454 | 2.01 |
ENSDART00000160123
ENSDART00000169653 |
pcdh2ab9
pcdh2ac
|
protocadherin 2 alpha b 9 protocadherin 2 alpha c |
| chr13_-_31470439 | 2.00 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr11_+_329687 | 1.96 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr19_+_1184878 | 1.82 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr20_+_48782068 | 1.80 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr23_-_44226556 | 1.79 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr5_-_22952156 | 1.72 |
ENSDART00000111146
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr1_-_21409877 | 1.67 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr16_+_20926673 | 1.67 |
ENSDART00000009827
|
hoxa2b
|
homeobox A2b |
| chr24_-_38816725 | 1.67 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr3_-_21280373 | 1.67 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr20_-_35246150 | 1.66 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr6_+_4539953 | 1.64 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr11_+_7324704 | 1.64 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr24_+_14713776 | 1.62 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr1_-_20911297 | 1.60 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr4_+_76608793 | 1.60 |
ENSDART00000141114
|
ms4a17a.8
|
membrane-spanning 4-domains, subfamily A, member 17A.8 |
| chr10_-_27223827 | 1.57 |
ENSDART00000185138
|
auts2a
|
autism susceptibility candidate 2a |
| chr21_+_51521 | 1.53 |
ENSDART00000007672
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr25_-_15040369 | 1.52 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr23_+_45611649 | 1.47 |
ENSDART00000169521
|
dclk2b
|
doublecortin-like kinase 2b |
| chr11_-_42099645 | 1.47 |
ENSDART00000173312
|
abhd6a
|
abhydrolase domain containing 6a |
| chr21_+_11468642 | 1.43 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr14_-_9199968 | 1.42 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr2_-_22688651 | 1.42 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr7_+_32722227 | 1.41 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr18_-_5692292 | 1.41 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr20_-_26531850 | 1.40 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr20_-_44557037 | 1.38 |
ENSDART00000140995
|
mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr18_+_19974289 | 1.38 |
ENSDART00000090334
ENSDART00000192982 |
skor1b
|
SKI family transcriptional corepressor 1b |
| chr15_-_23699737 | 1.38 |
ENSDART00000078311
|
zgc:154093
|
zgc:154093 |
| chr6_-_11780070 | 1.36 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr1_-_38709551 | 1.36 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr1_+_20635190 | 1.35 |
ENSDART00000145418
ENSDART00000148518 ENSDART00000139461 ENSDART00000102969 ENSDART00000166479 |
spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_-_13772847 | 1.33 |
ENSDART00000145103
ENSDART00000184491 |
cntfr
|
ciliary neurotrophic factor receptor |
| chr7_+_67451108 | 1.33 |
ENSDART00000163840
|
gcshb
|
glycine cleavage system protein H (aminomethyl carrier), b |
| chr17_+_53311618 | 1.31 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr17_-_52594756 | 1.28 |
ENSDART00000167288
|
si:ch211-173a9.7
|
si:ch211-173a9.7 |
| chr20_-_26532167 | 1.28 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr25_+_37209619 | 1.28 |
ENSDART00000112192
|
si:dkey-234i14.3
|
si:dkey-234i14.3 |
| chr7_+_73332486 | 1.26 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr11_-_33960318 | 1.22 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr22_-_5006801 | 1.21 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr4_-_5764255 | 1.21 |
ENSDART00000113864
|
faxca
|
failed axon connections homolog a |
| chr1_-_8428736 | 1.21 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr17_+_51764310 | 1.20 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr25_-_18140537 | 1.20 |
ENSDART00000113581
|
kitlga
|
kit ligand a |
| chr3_-_18575868 | 1.19 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr11_+_15931188 | 1.19 |
ENSDART00000165768
ENSDART00000081098 |
draxin
|
dorsal inhibitory axon guidance protein |
| chr13_-_5569562 | 1.19 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr7_+_39624728 | 1.18 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr25_+_3677650 | 1.18 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr23_+_45611980 | 1.17 |
ENSDART00000181582
|
dclk2b
|
doublecortin-like kinase 2b |
| chr25_+_14165447 | 1.17 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_-_50859053 | 1.17 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr7_+_48288762 | 1.16 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr5_+_63668735 | 1.14 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr8_+_48603398 | 1.14 |
ENSDART00000074900
|
zgc:195023
|
zgc:195023 |
| chr6_+_52350443 | 1.14 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr1_-_40911332 | 1.11 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr12_-_37299646 | 1.11 |
ENSDART00000146142
ENSDART00000085201 |
pmp22b
|
peripheral myelin protein 22b |
| chr24_+_10413484 | 1.11 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr7_+_73593814 | 1.09 |
ENSDART00000110544
|
znf219
|
zinc finger protein 219 |
| chr9_-_46842179 | 1.08 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr14_+_52369262 | 1.08 |
ENSDART00000169352
ENSDART00000157833 |
igfbp7
|
insulin-like growth factor binding protein 7 |
| chr5_+_28973264 | 1.07 |
ENSDART00000005638
|
stxbp1b
|
syntaxin binding protein 1b |
| chr25_+_21324588 | 1.07 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr23_+_45456490 | 1.07 |
ENSDART00000036631
|
cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr12_-_1951233 | 1.06 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr14_-_1454045 | 1.06 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr23_-_39849155 | 1.05 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr12_-_48477031 | 1.05 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr9_-_14504834 | 1.05 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr25_+_4837915 | 1.04 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr5_+_28972935 | 1.04 |
ENSDART00000193274
|
stxbp1b
|
syntaxin binding protein 1b |
| chr14_-_45206999 | 1.03 |
ENSDART00000110191
|
shisa3
|
shisa family member 3 |
| chr20_-_3953631 | 1.02 |
ENSDART00000124197
|
CU929133.1
|
|
| chr20_+_25350435 | 1.02 |
ENSDART00000063034
|
fam228a
|
family with sequence similarity 228, member A |
| chr22_-_29586608 | 1.02 |
ENSDART00000059869
|
adra2a
|
adrenoceptor alpha 2A |
| chr24_+_5840748 | 1.01 |
ENSDART00000139191
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr12_-_7607114 | 1.01 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr3_+_39099716 | 1.00 |
ENSDART00000083388
|
tmem98
|
transmembrane protein 98 |
| chr10_+_15064433 | 1.00 |
ENSDART00000179978
|
parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr18_+_62932 | 1.00 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr25_-_20381271 | 0.99 |
ENSDART00000142665
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr2_-_24462277 | 0.99 |
ENSDART00000033922
|
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr13_+_35746440 | 0.98 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr3_-_43356082 | 0.97 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr11_-_10828539 | 0.97 |
ENSDART00000040180
|
tbr1a
|
T-box, brain, 1a |
| chr24_-_28245872 | 0.96 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr1_-_36151377 | 0.96 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr15_+_36941490 | 0.96 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr10_+_45345574 | 0.95 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr13_-_40401870 | 0.93 |
ENSDART00000128951
|
nkx3.3
|
NK3 homeobox 3 |
| chr16_+_28754403 | 0.92 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr9_+_8929764 | 0.92 |
ENSDART00000102562
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr10_-_37075361 | 0.91 |
ENSDART00000132023
|
myo18aa
|
myosin XVIIIAa |
| chr20_+_26892761 | 0.91 |
ENSDART00000133293
|
ftr97
|
finTRIM family, member 97 |
| chr16_+_49601838 | 0.91 |
ENSDART00000168570
ENSDART00000159236 |
si:dkey-82o10.4
|
si:dkey-82o10.4 |
| chr13_+_30054996 | 0.90 |
ENSDART00000110061
ENSDART00000186045 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr17_+_38566717 | 0.89 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr25_-_4733008 | 0.86 |
ENSDART00000191884
|
drd4a
|
dopamine receptor D4a |
| chr7_-_52334429 | 0.86 |
ENSDART00000187372
|
CR938716.1
|
|
| chr20_-_1314537 | 0.86 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr24_+_4977862 | 0.84 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr22_-_21314821 | 0.84 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr13_-_31829786 | 0.84 |
ENSDART00000138667
|
sertad4
|
SERTA domain containing 4 |
| chr5_-_31712399 | 0.83 |
ENSDART00000141328
|
pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr5_-_21030934 | 0.83 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr16_+_32559821 | 0.83 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr23_-_32129569 | 0.83 |
ENSDART00000167761
ENSDART00000139569 |
zgc:92658
|
zgc:92658 |
| chr18_+_50627085 | 0.83 |
ENSDART00000161429
|
chrnb4
|
cholinergic receptor, nicotinic, beta 4 (neuronal) |
| chr11_+_575665 | 0.81 |
ENSDART00000122133
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr13_+_29773153 | 0.81 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr1_-_22834824 | 0.80 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr2_-_30324610 | 0.79 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr12_+_1398404 | 0.79 |
ENSDART00000026303
|
rasd1
|
RAS, dexamethasone-induced 1 |
| chr16_-_26296477 | 0.79 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr3_-_1204341 | 0.78 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr1_-_47771742 | 0.77 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr2_-_5399437 | 0.77 |
ENSDART00000132411
|
si:ch1073-184j22.2
|
si:ch1073-184j22.2 |
| chr6_+_56147812 | 0.77 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr24_+_4978055 | 0.75 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr6_+_120181 | 0.75 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr24_+_35881124 | 0.75 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr20_-_54869006 | 0.75 |
ENSDART00000184817
|
CABZ01037174.1
|
|
| chr11_-_29916607 | 0.74 |
ENSDART00000138912
ENSDART00000079152 |
vegfd
|
vascular endothelial growth factor D |
| chr3_-_36115339 | 0.74 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr1_+_9004719 | 0.73 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr3_+_36313532 | 0.73 |
ENSDART00000151305
|
slc16a6b
|
solute carrier family 16, member 6b |
| chr25_-_18140305 | 0.72 |
ENSDART00000180222
|
kitlga
|
kit ligand a |
| chr1_-_46984142 | 0.71 |
ENSDART00000125032
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr18_-_3166726 | 0.70 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr4_-_9810999 | 0.70 |
ENSDART00000146858
|
si:dkeyp-27e10.3
|
si:dkeyp-27e10.3 |
| chr13_+_27073901 | 0.70 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr23_+_26039524 | 0.69 |
ENSDART00000142851
|
itih6
|
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr10_+_37981471 | 0.68 |
ENSDART00000113112
ENSDART00000185102 |
wscd1b
|
WSC domain containing 1b |
| chr7_+_14632157 | 0.68 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr19_+_41075488 | 0.68 |
ENSDART00000138687
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr1_-_44161417 | 0.68 |
ENSDART00000083213
|
slc43a3a
|
solute carrier family 43, member 3a |
| chr3_+_17744339 | 0.68 |
ENSDART00000132622
|
znf385c
|
zinc finger protein 385C |
| chr8_+_23916647 | 0.68 |
ENSDART00000143152
|
cpne5a
|
copine Va |
| chr20_+_50852356 | 0.67 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr16_-_560574 | 0.67 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr7_+_24114694 | 0.66 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr24_+_5840258 | 0.66 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr13_-_29421331 | 0.66 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr2_-_30324297 | 0.66 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr7_+_4162994 | 0.65 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr14_+_11103718 | 0.65 |
ENSDART00000161311
|
nexmifb
|
neurite extension and migration factor b |
| chr20_-_1314355 | 0.64 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr4_+_3980247 | 0.64 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr7_-_49594995 | 0.64 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr9_+_53707240 | 0.64 |
ENSDART00000171490
|
PCDH8
|
si:ch211-199f5.1 |
| chr1_-_53700392 | 0.63 |
ENSDART00000127216
|
fam161a
|
family with sequence similarity 161, member A |
| chr2_-_44720551 | 0.62 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr22_-_12160283 | 0.62 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr18_+_48428126 | 0.62 |
ENSDART00000137978
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr9_+_6587056 | 0.62 |
ENSDART00000193421
|
fhl2a
|
four and a half LIM domains 2a |
| chr13_+_31757331 | 0.61 |
ENSDART00000044282
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr11_-_36765834 | 0.60 |
ENSDART00000002055
|
cacna2d3
|
calcium channel, voltage dependent, alpha2/delta subunit 3 |
| chr20_-_8419057 | 0.60 |
ENSDART00000145841
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr25_-_4733221 | 0.60 |
ENSDART00000172689
|
drd4a
|
dopamine receptor D4a |
| chr20_-_40119872 | 0.59 |
ENSDART00000191569
|
nkain2
|
sodium/potassium transporting ATPase interacting 2 |
| chr24_+_35881984 | 0.59 |
ENSDART00000066587
|
klhl14
|
kelch-like family member 14 |
| chr22_-_29922872 | 0.58 |
ENSDART00000020249
|
dusp5
|
dual specificity phosphatase 5 |
| chr25_-_3979583 | 0.58 |
ENSDART00000124749
|
myrf
|
myelin regulatory factor |
| chr7_-_37858569 | 0.57 |
ENSDART00000129156
ENSDART00000173552 ENSDART00000178306 |
adcy7
|
adenylate cyclase 7 |
| chr3_-_18410968 | 0.56 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr20_-_9963713 | 0.56 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr23_+_44644911 | 0.56 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr13_+_111212 | 0.56 |
ENSDART00000167840
|
dnaaf2
|
dynein, axonemal, assembly factor 2 |
| chr20_+_28245164 | 0.55 |
ENSDART00000103320
|
dll4
|
delta-like 4 (Drosophila) |
| chr22_-_7006974 | 0.55 |
ENSDART00000133143
|
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.7 | 2.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.5 | 2.4 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 2.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 2.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 1.7 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.3 | 1.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.3 | 1.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 0.9 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.2 | 1.4 | GO:0006545 | glycine metabolic process(GO:0006544) glycine biosynthetic process(GO:0006545) |
| 0.2 | 1.2 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.2 | 1.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 0.7 | GO:0072579 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 2.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 0.7 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.2 | 1.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 3.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 1.0 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 1.3 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.2 | 0.9 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 1.0 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.2 | 0.6 | GO:2000048 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 1.0 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.2 | 2.8 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 2.0 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.2 | 1.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 1.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 2.7 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.4 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.1 | 18.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.4 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.5 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 1.8 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 0.4 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 4.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 2.0 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 1.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 0.8 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.4 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 1.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 1.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.8 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 1.1 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.6 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 1.2 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.2 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.7 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.3 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.7 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.1 | 3.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.3 | GO:0099623 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 1.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 2.2 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.5 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.1 | 1.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 1.7 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 0.2 | GO:0048898 | anterior lateral line system development(GO:0048898) |
| 0.1 | 2.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.5 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 0.3 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 2.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 1.2 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 1.0 | GO:0039022 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
| 0.0 | 0.7 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.0 | 1.0 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.1 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.8 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 1.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.3 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 1.9 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) negative regulation of intracellular protein transport(GO:0090317) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 2.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 1.1 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.6 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.6 | GO:0071453 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 0.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 1.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.3 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.6 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.8 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.5 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.3 | 1.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 2.7 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 3.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 0.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 2.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 3.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.4 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.3 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 2.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.9 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 0.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0044306 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.5 | 2.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 2.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.4 | 2.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 1.5 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.3 | 1.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.7 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.2 | 1.4 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 18.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.7 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 2.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 1.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 1.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 3.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 1.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 3.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 1.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 4.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 2.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 2.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.0 | 0.3 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 2.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 1.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 2.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 17.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 4.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.8 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 2.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.5 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |