Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
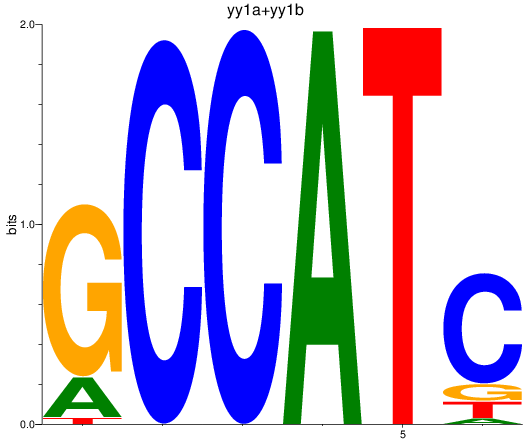
Results for yy1a+yy1b
Z-value: 1.15
Transcription factors associated with yy1a+yy1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
yy1b
|
ENSDARG00000027978 | YY1 transcription factor b |
|
yy1a
|
ENSDARG00000042796 | YY1 transcription factor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| yy1a | dr11_v1_chr17_-_30863252_30863252 | -0.65 | 6.0e-02 | Click! |
| yy1b | dr11_v1_chr20_-_54435287_54435287 | 0.34 | 3.7e-01 | Click! |
Activity profile of yy1a+yy1b motif
Sorted Z-values of yy1a+yy1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_35892767 | 0.82 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr22_+_24318131 | 0.79 |
ENSDART00000187360
ENSDART00000165618 |
ccdc50
|
coiled-coil domain containing 50 |
| chr15_-_35126332 | 0.78 |
ENSDART00000007636
|
zgc:55413
|
zgc:55413 |
| chr1_-_18811517 | 0.71 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr2_+_24638367 | 0.68 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr14_-_41468892 | 0.68 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr12_+_20693743 | 0.67 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr10_+_44057502 | 0.64 |
ENSDART00000183868
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr8_+_36503797 | 0.64 |
ENSDART00000184785
|
slc7a4
|
solute carrier family 7, member 4 |
| chr16_+_48714048 | 0.63 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr8_+_3434146 | 0.63 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr12_+_21525496 | 0.60 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr10_+_37182626 | 0.59 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr15_+_19885498 | 0.57 |
ENSDART00000152123
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr21_-_13690712 | 0.57 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr18_-_12654011 | 0.56 |
ENSDART00000062343
|
hipk2
|
homeodomain interacting protein kinase 2 |
| chr23_-_44577885 | 0.56 |
ENSDART00000166654
|
si:ch73-160p18.4
|
si:ch73-160p18.4 |
| chr21_-_34261677 | 0.56 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr9_+_2393764 | 0.56 |
ENSDART00000172624
|
chn1
|
chimerin 1 |
| chr16_-_8120203 | 0.55 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr14_-_30932373 | 0.55 |
ENSDART00000172988
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr4_+_23223881 | 0.53 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr18_-_43866001 | 0.53 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr17_+_52300018 | 0.53 |
ENSDART00000190302
|
esrrb
|
estrogen-related receptor beta |
| chr10_+_44057177 | 0.52 |
ENSDART00000164610
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr1_+_218524 | 0.52 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr5_-_9678075 | 0.51 |
ENSDART00000097217
|
FAM109B
|
si:ch211-193c2.2 |
| chr4_-_15003854 | 0.51 |
ENSDART00000134701
ENSDART00000002401 |
klhdc10
|
kelch domain containing 10 |
| chr9_-_32142311 | 0.50 |
ENSDART00000142768
|
ankrd44
|
ankyrin repeat domain 44 |
| chr13_+_28618086 | 0.50 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr20_-_15161502 | 0.49 |
ENSDART00000187072
|
plpp6
|
phospholipid phosphatase 6 |
| chr6_-_9565526 | 0.49 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr11_+_18053333 | 0.48 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr14_-_46617228 | 0.47 |
ENSDART00000161010
ENSDART00000171054 |
prom1a
|
prominin 1a |
| chr15_-_21669618 | 0.47 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr11_+_18130300 | 0.46 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr11_+_18157260 | 0.46 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr5_+_27583117 | 0.46 |
ENSDART00000180340
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr12_+_19976400 | 0.45 |
ENSDART00000153177
|
mkl2a
|
MKL/myocardin-like 2a |
| chr9_+_13641668 | 0.45 |
ENSDART00000135877
|
als2a
|
amyotrophic lateral sclerosis 2a (juvenile) |
| chr5_+_24047292 | 0.45 |
ENSDART00000029889
|
ctdnep1a
|
CTD nuclear envelope phosphatase 1a |
| chr3_-_2613990 | 0.45 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr5_+_27583445 | 0.45 |
ENSDART00000136488
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr11_+_18037729 | 0.44 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr17_+_33158350 | 0.44 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr18_+_14307059 | 0.44 |
ENSDART00000186558
|
zgc:173742
|
zgc:173742 |
| chr25_-_7670391 | 0.44 |
ENSDART00000044970
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr5_-_13848296 | 0.44 |
ENSDART00000127109
|
figla
|
folliculogenesis specific bHLH transcription factor |
| chr15_-_25613114 | 0.44 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr19_+_8612839 | 0.44 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr15_-_33304133 | 0.43 |
ENSDART00000186092
|
nbeab
|
neurobeachin b |
| chr18_+_15106518 | 0.43 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr13_+_16522608 | 0.43 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr25_+_33063762 | 0.43 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr22_-_20838011 | 0.43 |
ENSDART00000186056
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr8_-_12847483 | 0.43 |
ENSDART00000146186
|
si:dkey-104n9.1
|
si:dkey-104n9.1 |
| chr7_+_44802353 | 0.42 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr20_+_23501535 | 0.42 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr25_+_5604512 | 0.42 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr7_-_4107423 | 0.41 |
ENSDART00000172971
|
zgc:55733
|
zgc:55733 |
| chr17_+_43863708 | 0.41 |
ENSDART00000133874
ENSDART00000140316 ENSDART00000142929 ENSDART00000148090 |
zgc:66313
|
zgc:66313 |
| chr10_+_15608326 | 0.41 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr25_+_28282274 | 0.41 |
ENSDART00000164502
|
aass
|
aminoadipate-semialdehyde synthase |
| chr3_+_33745014 | 0.41 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr8_-_49207319 | 0.41 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr5_-_48664522 | 0.41 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr21_+_43199237 | 0.41 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr17_-_51829310 | 0.40 |
ENSDART00000154544
|
numb
|
numb homolog (Drosophila) |
| chr5_-_37875636 | 0.40 |
ENSDART00000184674
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr3_-_10612186 | 0.40 |
ENSDART00000191034
|
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr17_+_24597001 | 0.40 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr16_-_33076764 | 0.40 |
ENSDART00000192917
|
dopey1
|
dopey family member 1 |
| chr10_-_26202766 | 0.40 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr6_+_4387150 | 0.39 |
ENSDART00000181283
|
rbm26
|
RNA binding motif protein 26 |
| chr3_+_43102010 | 0.39 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr2_-_21621878 | 0.39 |
ENSDART00000189540
|
zgc:55781
|
zgc:55781 |
| chr20_-_31743553 | 0.39 |
ENSDART00000087405
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr10_+_2234283 | 0.38 |
ENSDART00000136363
|
cntnap3
|
contactin associated protein like 3 |
| chr25_-_9805269 | 0.38 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr10_-_33251876 | 0.38 |
ENSDART00000184565
|
bcl7ba
|
BCL tumor suppressor 7Ba |
| chr18_+_44631789 | 0.38 |
ENSDART00000144271
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr2_-_31686353 | 0.38 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr14_+_31529958 | 0.38 |
ENSDART00000053026
|
fam122b
|
family with sequence similarity 122B |
| chr6_+_19950107 | 0.37 |
ENSDART00000181632
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr10_-_34002185 | 0.37 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr8_-_1730686 | 0.37 |
ENSDART00000183802
|
CABZ01065416.1
|
|
| chr11_-_2270069 | 0.37 |
ENSDART00000189005
|
znf740a
|
zinc finger protein 740a |
| chr1_-_40519340 | 0.36 |
ENSDART00000114659
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr22_+_17359346 | 0.36 |
ENSDART00000145434
|
gpr52
|
G protein-coupled receptor 52 |
| chr21_+_3897680 | 0.36 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr3_+_17456428 | 0.36 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr19_+_29303847 | 0.36 |
ENSDART00000009149
|
maco1a
|
macoilin 1a |
| chr3_-_21118969 | 0.36 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr20_-_52928541 | 0.36 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr23_-_29878643 | 0.36 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr8_+_9699111 | 0.36 |
ENSDART00000111853
|
gripap1
|
GRIP1 associated protein 1 |
| chr13_-_42066299 | 0.36 |
ENSDART00000111536
|
rmdn2
|
regulator of microtubule dynamics 2 |
| chr17_-_5732299 | 0.36 |
ENSDART00000017244
|
xkr6a
|
XK, Kell blood group complex subunit-related family, member 6a |
| chr20_-_15161669 | 0.36 |
ENSDART00000080333
ENSDART00000063882 |
plpp6
|
phospholipid phosphatase 6 |
| chr18_+_173603 | 0.36 |
ENSDART00000185918
|
larp6a
|
La ribonucleoprotein domain family, member 6a |
| chr19_-_20403507 | 0.35 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr3_-_44753325 | 0.35 |
ENSDART00000074543
|
hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr8_-_54077740 | 0.35 |
ENSDART00000027000
|
rho
|
rhodopsin |
| chr23_-_30431333 | 0.35 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr12_+_27285994 | 0.34 |
ENSDART00000087204
|
dusp3a
|
dual specificity phosphatase 3a |
| chr10_+_14982977 | 0.34 |
ENSDART00000140869
|
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr19_-_20403845 | 0.34 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr2_+_50862527 | 0.34 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr3_+_4997545 | 0.34 |
ENSDART00000181237
|
CABZ01117706.1
|
|
| chr24_+_26379441 | 0.34 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr12_-_13729263 | 0.34 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr23_+_25172682 | 0.34 |
ENSDART00000191197
ENSDART00000183497 |
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr15_-_15965456 | 0.33 |
ENSDART00000154579
|
synrg
|
synergin, gamma |
| chr7_+_50109239 | 0.33 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr6_+_1724889 | 0.33 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr15_+_24005289 | 0.33 |
ENSDART00000088648
|
myo18ab
|
myosin XVIIIAb |
| chr5_+_24520437 | 0.33 |
ENSDART00000144560
|
dgcr8
|
DGCR8 microprocessor complex subunit |
| chr13_-_22903246 | 0.33 |
ENSDART00000089133
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr6_-_31576397 | 0.33 |
ENSDART00000111837
|
raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr6_-_51386656 | 0.33 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr12_-_35787801 | 0.33 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr18_+_3634652 | 0.33 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr8_+_8671229 | 0.33 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr23_-_27501923 | 0.33 |
ENSDART00000188394
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr3_+_14571813 | 0.33 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr12_-_10508952 | 0.33 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr21_-_40557281 | 0.33 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr14_+_1240419 | 0.33 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr6_-_7842078 | 0.33 |
ENSDART00000065507
|
plppr2b
|
phospholipid phosphatase related 2b |
| chr5_+_29851433 | 0.33 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr11_+_25472758 | 0.33 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr20_+_23670494 | 0.33 |
ENSDART00000155262
|
nek1
|
NIMA-related kinase 1 |
| chr19_+_32456974 | 0.33 |
ENSDART00000088265
|
atxn1a
|
ataxin 1a |
| chr2_+_30032303 | 0.32 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr5_+_29652513 | 0.32 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr20_+_35445462 | 0.32 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr24_+_14937205 | 0.32 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr2_+_24700922 | 0.32 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr5_+_42912966 | 0.32 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr4_-_21584158 | 0.32 |
ENSDART00000132923
|
ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr12_-_41759686 | 0.32 |
ENSDART00000172175
ENSDART00000165152 |
ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr23_-_24483311 | 0.32 |
ENSDART00000185793
ENSDART00000109248 |
spen
|
spen family transcriptional repressor |
| chr12_-_7234915 | 0.32 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr22_-_16377666 | 0.31 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr7_-_4461104 | 0.31 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr1_-_41374117 | 0.31 |
ENSDART00000074777
|
htt
|
huntingtin |
| chr17_-_25331439 | 0.31 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr23_-_32390742 | 0.31 |
ENSDART00000032861
|
ankrd52a
|
ankyrin repeat domain 52a |
| chr3_-_1263047 | 0.31 |
ENSDART00000184388
|
tcf20
|
transcription factor 20 |
| chr15_+_23951560 | 0.31 |
ENSDART00000191133
|
myo18ab
|
myosin XVIIIAb |
| chr11_-_25461336 | 0.31 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr4_-_277081 | 0.31 |
ENSDART00000166174
|
si:ch73-252i11.1
|
si:ch73-252i11.1 |
| chr18_+_660578 | 0.31 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr7_-_55292116 | 0.31 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr7_-_69795488 | 0.31 |
ENSDART00000162414
|
USP53 (1 of many)
|
ubiquitin specific peptidase 53 |
| chr20_-_18535502 | 0.31 |
ENSDART00000049437
|
cdc42bpb
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr21_-_30026359 | 0.31 |
ENSDART00000153645
|
pwwp2a
|
PWWP domain containing 2A |
| chr21_+_37513488 | 0.31 |
ENSDART00000185394
|
amot
|
angiomotin |
| chr8_-_32354677 | 0.31 |
ENSDART00000138268
ENSDART00000133245 ENSDART00000179677 ENSDART00000174450 |
ipo11
|
importin 11 |
| chr2_-_10631767 | 0.30 |
ENSDART00000190033
|
mtf2
|
metal response element binding transcription factor 2 |
| chr8_-_14604606 | 0.30 |
ENSDART00000090254
ENSDART00000188953 |
cep350
|
centrosomal protein 350 |
| chr15_-_28907709 | 0.30 |
ENSDART00000017268
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr24_+_10202718 | 0.30 |
ENSDART00000126668
|
pou6f2
|
POU class 6 homeobox 2 |
| chr8_-_40442979 | 0.30 |
ENSDART00000157698
ENSDART00000192657 |
tbc1d10aa
|
TBC1 domain family, member 10Aa |
| chr15_-_28908027 | 0.30 |
ENSDART00000182790
ENSDART00000192461 |
eml2
|
echinoderm microtubule associated protein like 2 |
| chr5_-_57311037 | 0.30 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr25_-_37084032 | 0.30 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr14_-_51619388 | 0.30 |
ENSDART00000170949
|
unc5a
|
unc-5 netrin receptor A |
| chr5_+_36768674 | 0.30 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr20_+_18260358 | 0.30 |
ENSDART00000187734
ENSDART00000191947 ENSDART00000057039 |
taf4b
|
TAF4B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr16_-_42965192 | 0.30 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr20_-_44576949 | 0.30 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr15_-_28904371 | 0.30 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr15_-_47107557 | 0.30 |
ENSDART00000111880
|
CABZ01079080.1
|
|
| chr4_-_23643272 | 0.30 |
ENSDART00000112301
ENSDART00000133184 |
trhde.2
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 2 |
| chr17_+_19626479 | 0.30 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr24_+_40905100 | 0.30 |
ENSDART00000167854
|
scn12ab
|
sodium channel, voltage gated, type XII, alpha b |
| chr17_-_27223965 | 0.29 |
ENSDART00000192577
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr9_-_12034444 | 0.29 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr3_-_21166597 | 0.29 |
ENSDART00000175941
|
taok2a
|
TAO kinase 2a |
| chr15_+_19991280 | 0.29 |
ENSDART00000186677
|
zgc:112083
|
zgc:112083 |
| chr5_-_31773208 | 0.29 |
ENSDART00000137556
ENSDART00000122066 |
fam102ab
|
family with sequence similarity 102, member Ab |
| chr9_+_33154841 | 0.29 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr3_+_3519607 | 0.29 |
ENSDART00000150922
|
cdc42ep1b
|
CDC42 effector protein (Rho GTPase binding) 1b |
| chr13_-_51247529 | 0.29 |
ENSDART00000191774
ENSDART00000083788 |
AL929217.1
|
|
| chr19_-_20403318 | 0.29 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr17_-_17447899 | 0.29 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr7_-_61279523 | 0.29 |
ENSDART00000098610
|
rell1
|
RELT like 1 |
| chr16_-_24642814 | 0.29 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr22_+_26703026 | 0.29 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr12_-_31484677 | 0.29 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr9_-_14137295 | 0.29 |
ENSDART00000127640
ENSDART00000189018 ENSDART00000188985 |
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr11_+_30817943 | 0.29 |
ENSDART00000150130
ENSDART00000159997 |
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr9_-_32804953 | 0.29 |
ENSDART00000134763
|
zgc:112056
|
zgc:112056 |
| chr22_-_16377960 | 0.29 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr17_+_20504196 | 0.29 |
ENSDART00000190539
|
neurl1ab
|
neuralized E3 ubiquitin protein ligase 1Ab |
| chr7_+_69449814 | 0.28 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr9_+_38502524 | 0.28 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr21_+_34132747 | 0.28 |
ENSDART00000148115
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr5_-_18513950 | 0.28 |
ENSDART00000145878
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of yy1a+yy1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.2 | 0.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 0.5 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 1.0 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.7 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.6 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.4 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.1 | GO:0035552 | oxidative DNA demethylation(GO:0035511) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 0.4 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.3 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 0.4 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.3 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.4 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.3 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 0.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.4 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.7 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 0.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.6 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.9 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 0.2 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.1 | 0.8 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.3 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.6 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 1.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.3 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.4 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.1 | 0.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.5 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.3 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.2 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.3 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.3 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.2 | GO:0099542 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism metabolic process(GO:0044033) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.4 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 1.4 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.4 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.0 | 0.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.6 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:0016038 | light absorption(GO:0016037) absorption of visible light(GO:0016038) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.3 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.2 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.2 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.5 | GO:0007350 | blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.2 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.3 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:0036088 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.1 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.3 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.4 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.4 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.0 | 0.3 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.3 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.1 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.1 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.6 | GO:0060415 | muscle tissue morphogenesis(GO:0060415) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.7 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0045162 | neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.1 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.4 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.8 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.7 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.9 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.0 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.2 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:0032602 | chemokine production(GO:0032602) thymocyte migration(GO:0072679) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.2 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0099640 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0030643 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.1 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.0 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.2 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 1.0 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.7 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.3 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0097346 | Swr1 complex(GO:0000812) INO80-type complex(GO:0097346) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.6 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.0 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.2 | 0.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.3 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.3 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 0.2 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.1 | 0.3 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.2 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 1.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.2 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.2 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.6 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.6 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.0 | 0.4 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.0 | 0.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.8 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 2.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0043548 | insulin receptor binding(GO:0005158) phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 1.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.0 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 2.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME SIGNALLING TO RAS | Genes involved in Signalling to RAS |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.5 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |