Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
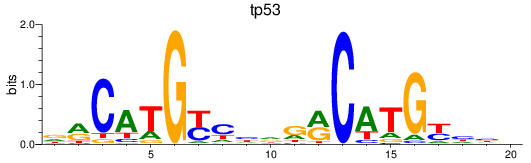
Results for tp53
Z-value: 0.98
Transcription factors associated with tp53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tp53
|
ENSDARG00000035559 | tumor protein p53 |
|
tp53
|
ENSDARG00000115148 | tumor protein p53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tp53 | dr11_v1_chr5_+_24089334_24089334 | 0.46 | 2.1e-01 | Click! |
Activity profile of tp53 motif
Sorted Z-values of tp53 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_35043007 | 1.67 |
ENSDART00000086954
|
sesn3
|
sestrin 3 |
| chr24_+_22485710 | 1.28 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr21_+_25688388 | 1.17 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr20_-_27225876 | 1.16 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr25_-_7520937 | 1.15 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr10_-_4980150 | 1.11 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr15_-_29387446 | 1.08 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr23_-_5759242 | 0.99 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr19_-_27462720 | 0.99 |
ENSDART00000135901
|
si:ch73-25f10.6
|
si:ch73-25f10.6 |
| chr22_+_37888249 | 0.91 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr10_+_8629275 | 0.84 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr23_+_23232136 | 0.84 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr25_+_34641536 | 0.75 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr10_+_4987766 | 0.71 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr6_+_49095646 | 0.69 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr4_+_25630555 | 0.67 |
ENSDART00000133425
|
acot15
|
acyl-CoA thioesterase 15 |
| chr5_-_57686487 | 0.66 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr15_-_18067220 | 0.66 |
ENSDART00000113142
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr14_+_36231126 | 0.65 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr23_-_31428763 | 0.62 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr10_+_38512270 | 0.57 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr18_+_45200043 | 0.56 |
ENSDART00000015786
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr4_-_40589466 | 0.55 |
ENSDART00000133295
ENSDART00000124872 |
si:dkey-207m2.4
|
si:dkey-207m2.4 |
| chr18_-_16924221 | 0.53 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr5_+_27898226 | 0.51 |
ENSDART00000098604
ENSDART00000180251 |
adam28
|
ADAM metallopeptidase domain 28 |
| chr12_+_27537357 | 0.51 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr13_+_28705143 | 0.51 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr7_+_26049818 | 0.49 |
ENSDART00000173611
|
si:dkey-6n21.12
|
si:dkey-6n21.12 |
| chr7_-_26049282 | 0.48 |
ENSDART00000136389
ENSDART00000101124 |
rnaseka
|
ribonuclease, RNase K a |
| chr8_+_28593707 | 0.48 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr9_+_51882534 | 0.47 |
ENSDART00000165493
|
CABZ01079550.1
|
|
| chr3_-_58205045 | 0.47 |
ENSDART00000130740
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr20_-_32332736 | 0.46 |
ENSDART00000062982
|
foxo3b
|
forkhead box O3b |
| chr3_-_50277959 | 0.46 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr11_+_6281647 | 0.46 |
ENSDART00000002459
|
ctns
|
cystinosin, lysosomal cystine transporter |
| chr9_-_15789526 | 0.44 |
ENSDART00000141318
|
si:dkey-103d23.3
|
si:dkey-103d23.3 |
| chr2_-_27900518 | 0.44 |
ENSDART00000109561
ENSDART00000077720 |
zgc:163121
|
zgc:163121 |
| chr4_+_23127104 | 0.44 |
ENSDART00000139543
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr19_+_34274504 | 0.44 |
ENSDART00000132046
|
si:ch211-9n13.3
|
si:ch211-9n13.3 |
| chr8_+_7322465 | 0.44 |
ENSDART00000192380
|
ssuh2rs1
|
ssu-2 homolog, related sequence 1 |
| chr7_+_48805534 | 0.42 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr23_-_1017428 | 0.41 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr15_+_627619 | 0.40 |
ENSDART00000157207
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr6_-_3514529 | 0.40 |
ENSDART00000151020
ENSDART00000082151 |
uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr23_-_1017605 | 0.40 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr15_-_23761580 | 0.39 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr3_+_23692462 | 0.39 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr3_+_16229911 | 0.38 |
ENSDART00000121728
|
rpl19
|
ribosomal protein L19 |
| chr6_-_3514753 | 0.37 |
ENSDART00000151765
|
uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr3_-_47294365 | 0.37 |
ENSDART00000191039
|
tnfsf18
|
TNF superfamily member 18 |
| chr4_+_25607743 | 0.36 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr4_+_34417403 | 0.36 |
ENSDART00000186032
ENSDART00000167909 |
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr17_-_23412705 | 0.36 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr13_+_31144305 | 0.35 |
ENSDART00000189602
|
CR931802.4
|
|
| chr14_+_22132896 | 0.35 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr4_-_71311438 | 0.34 |
ENSDART00000168021
|
si:dkeyp-123a12.2
|
si:dkeyp-123a12.2 |
| chr5_-_6508250 | 0.32 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr20_+_52546186 | 0.32 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr19_+_9150041 | 0.32 |
ENSDART00000127803
ENSDART00000091533 |
clk2a
|
CDC-like kinase 2a |
| chr16_-_48400639 | 0.31 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr3_-_50146854 | 0.30 |
ENSDART00000157047
|
si:dkey-120c6.5
|
si:dkey-120c6.5 |
| chr16_-_30931651 | 0.29 |
ENSDART00000132643
|
slc45a4
|
solute carrier family 45, member 4 |
| chr5_-_29570141 | 0.28 |
ENSDART00000043259
|
entpd2a.2
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 2 |
| chr25_-_36044583 | 0.28 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr3_-_49815223 | 0.28 |
ENSDART00000181208
|
gcgra
|
glucagon receptor a |
| chr25_+_19095231 | 0.27 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr4_-_62206968 | 0.27 |
ENSDART00000193074
|
si:dkeyp-35e5.5
|
si:dkeyp-35e5.5 |
| chr3_-_31078348 | 0.27 |
ENSDART00000055360
|
elob
|
elongin B |
| chr3_-_11040575 | 0.27 |
ENSDART00000159802
|
CR382337.2
|
|
| chr20_-_26929685 | 0.25 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr15_+_44053244 | 0.25 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr19_+_43780970 | 0.24 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr16_-_30931468 | 0.24 |
ENSDART00000187838
|
slc45a4
|
solute carrier family 45, member 4 |
| chr16_+_35595312 | 0.24 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr5_+_24089334 | 0.24 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr9_+_29671553 | 0.24 |
ENSDART00000015377
|
rnaseh2b
|
ribonuclease H2, subunit B |
| chr5_-_69804433 | 0.24 |
ENSDART00000028954
|
rilpl2
|
Rab interacting lysosomal protein-like 2 |
| chr6_+_59913147 | 0.23 |
ENSDART00000156543
|
aamp
|
angio-associated, migratory cell protein |
| chr18_+_11970987 | 0.23 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr4_+_23127284 | 0.23 |
ENSDART00000122675
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr25_-_36044294 | 0.23 |
ENSDART00000182207
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr9_+_8929764 | 0.23 |
ENSDART00000102562
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr4_+_77808579 | 0.22 |
ENSDART00000166325
|
si:dkey-238k10.3
|
si:dkey-238k10.3 |
| chr4_-_70050522 | 0.22 |
ENSDART00000100252
|
si:ch211-171l17.14
|
si:ch211-171l17.14 |
| chr9_+_50110763 | 0.22 |
ENSDART00000162990
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr14_+_28438947 | 0.22 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr19_+_44009473 | 0.21 |
ENSDART00000151004
|
nkd3
|
naked cuticle homolog 3 |
| chr20_-_15132151 | 0.21 |
ENSDART00000063884
|
si:dkey-239i20.4
|
si:dkey-239i20.4 |
| chr10_-_27566481 | 0.21 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr8_+_23639124 | 0.20 |
ENSDART00000083108
|
nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr9_+_55075526 | 0.20 |
ENSDART00000139555
ENSDART00000133864 |
gpr143
|
G protein-coupled receptor 143 |
| chr7_+_4194055 | 0.20 |
ENSDART00000105073
|
si:dkey-28d5.14
|
si:dkey-28d5.14 |
| chr17_-_8268406 | 0.20 |
ENSDART00000149873
ENSDART00000064668 ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr17_+_25849332 | 0.19 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr13_+_1726953 | 0.19 |
ENSDART00000103004
|
zmp:0000000760
|
zmp:0000000760 |
| chr4_-_2440175 | 0.19 |
ENSDART00000058059
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr4_+_23126558 | 0.19 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr15_+_29113616 | 0.18 |
ENSDART00000190015
ENSDART00000191099 |
capns1b
|
calpain, small subunit 1 b |
| chr19_+_25154066 | 0.18 |
ENSDART00000163220
|
si:ch211-239d6.2
|
si:ch211-239d6.2 |
| chr17_-_22137199 | 0.18 |
ENSDART00000089689
|
mrps5
|
mitochondrial ribosomal protein S5 |
| chr25_-_35136267 | 0.18 |
ENSDART00000109751
|
zgc:173585
|
zgc:173585 |
| chr4_+_13931578 | 0.18 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr22_+_1947494 | 0.17 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr20_-_7133546 | 0.17 |
ENSDART00000160910
|
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr14_+_4796168 | 0.17 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr4_+_13931733 | 0.16 |
ENSDART00000141742
ENSDART00000067175 |
pphln1
|
periphilin 1 |
| chr18_+_5172848 | 0.16 |
ENSDART00000190642
|
CABZ01080599.1
|
|
| chr4_+_40265583 | 0.16 |
ENSDART00000151870
|
si:ch211-246n15.2
|
si:ch211-246n15.2 |
| chr21_+_5932140 | 0.16 |
ENSDART00000193767
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr4_+_34417798 | 0.16 |
ENSDART00000157757
|
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr4_+_77839900 | 0.15 |
ENSDART00000162243
|
si:dkey-207m2.4
|
si:dkey-207m2.4 |
| chr23_+_11285662 | 0.15 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr3_-_50147160 | 0.15 |
ENSDART00000191341
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr20_-_45708962 | 0.15 |
ENSDART00000124283
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr23_-_15090782 | 0.15 |
ENSDART00000133624
|
si:ch211-218g4.2
|
si:ch211-218g4.2 |
| chr8_-_48770603 | 0.14 |
ENSDART00000159712
|
pimr184
|
Pim proto-oncogene, serine/threonine kinase, related 184 |
| chr10_+_36029537 | 0.14 |
ENSDART00000165386
|
hmgb1a
|
high mobility group box 1a |
| chr17_-_10838434 | 0.14 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr3_+_30922947 | 0.14 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr20_-_43663494 | 0.14 |
ENSDART00000144564
|
BX470188.1
|
|
| chr4_+_59246094 | 0.14 |
ENSDART00000160587
|
si:dkey-105i14.1
|
si:dkey-105i14.1 |
| chr20_-_13673566 | 0.13 |
ENSDART00000181641
|
TAGAP
|
si:ch211-122h15.4 |
| chr13_+_4225173 | 0.13 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr4_-_13931508 | 0.13 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr11_-_13618412 | 0.13 |
ENSDART00000137289
|
sid1
|
secreted immunoglobulin domain 1 |
| chr22_+_9793923 | 0.13 |
ENSDART00000063323
|
si:dkey-102c8.2
|
si:dkey-102c8.2 |
| chr15_+_1134870 | 0.12 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr11_-_37693019 | 0.11 |
ENSDART00000102898
|
zgc:158258
|
zgc:158258 |
| chr9_+_56103173 | 0.11 |
ENSDART00000172063
|
edar
|
ectodysplasin A receptor |
| chr11_+_577092 | 0.11 |
ENSDART00000191827
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr14_+_46287296 | 0.11 |
ENSDART00000183620
|
cabp2b
|
calcium binding protein 2b |
| chr3_-_34082283 | 0.11 |
ENSDART00000151264
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr18_+_14619544 | 0.11 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr2_-_51316187 | 0.11 |
ENSDART00000163362
ENSDART00000166871 ENSDART00000160262 |
pigrl3.5
|
polymeric immunoglobulin receptor-like 3.5 |
| chr23_+_11347313 | 0.11 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr23_-_24047054 | 0.11 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr4_+_59245846 | 0.11 |
ENSDART00000104921
|
si:dkey-105i14.1
|
si:dkey-105i14.1 |
| chr20_-_26936887 | 0.10 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr6_-_18960105 | 0.10 |
ENSDART00000185278
ENSDART00000162968 |
sept9b
|
septin 9b |
| chr13_-_37189560 | 0.10 |
ENSDART00000134789
|
si:dkeyp-77c8.2
|
si:dkeyp-77c8.2 |
| chr10_+_35953068 | 0.10 |
ENSDART00000015279
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr4_-_13931293 | 0.10 |
ENSDART00000067172
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr16_+_40508882 | 0.09 |
ENSDART00000126129
|
nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr3_+_3992590 | 0.09 |
ENSDART00000180211
ENSDART00000054840 |
BX005421.2
|
|
| chr22_+_9854088 | 0.09 |
ENSDART00000122678
|
BX539307.1
|
|
| chr15_+_29362714 | 0.09 |
ENSDART00000099916
ENSDART00000126196 |
gucy2f
|
guanylate cyclase 2F, retinal |
| chr11_-_29737088 | 0.09 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr6_-_42049350 | 0.08 |
ENSDART00000022949
|
parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr20_+_7398090 | 0.08 |
ENSDART00000108649
|
pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr20_-_16404014 | 0.08 |
ENSDART00000152328
|
LRRC52
|
si:dkey-52j6.3 |
| chr8_+_15277874 | 0.08 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr7_-_20464133 | 0.08 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr14_+_36869273 | 0.08 |
ENSDART00000111674
|
F2RL3
|
si:ch211-132p1.2 |
| chr4_-_44509477 | 0.08 |
ENSDART00000102949
ENSDART00000135262 |
si:dkeyp-100h4.1
|
si:dkeyp-100h4.1 |
| chr7_-_2004159 | 0.07 |
ENSDART00000181965
ENSDART00000173821 |
zgc:171424
zgc:171424
|
zgc:171424 zgc:171424 |
| chr4_-_34455848 | 0.07 |
ENSDART00000184208
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr8_-_4694458 | 0.07 |
ENSDART00000019828
|
zgc:63587
|
zgc:63587 |
| chr5_+_29831235 | 0.06 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr10_+_35952532 | 0.06 |
ENSDART00000184730
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr5_-_1999417 | 0.06 |
ENSDART00000155437
ENSDART00000145781 |
si:ch211-160e1.5
|
si:ch211-160e1.5 |
| chr24_-_30862168 | 0.06 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr3_-_36602069 | 0.06 |
ENSDART00000165414
|
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr4_+_34418211 | 0.06 |
ENSDART00000160070
|
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr18_+_33340868 | 0.06 |
ENSDART00000099139
|
v2rh14
|
vomeronasal 2 receptor, h14 |
| chr1_-_11596829 | 0.06 |
ENSDART00000140725
|
si:dkey-26i13.7
|
si:dkey-26i13.7 |
| chr12_+_6098713 | 0.06 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr23_-_33775145 | 0.06 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr22_+_24770744 | 0.06 |
ENSDART00000142882
|
si:rp71-23d18.4
|
si:rp71-23d18.4 |
| chr3_-_7940522 | 0.06 |
ENSDART00000159710
ENSDART00000167201 |
trim35-25
|
tripartite motif containing 35-25 |
| chr14_+_16022157 | 0.06 |
ENSDART00000167092
|
npy7r
|
neuropeptide Y receptor Y7 |
| chr18_+_7150144 | 0.06 |
ENSDART00000114690
|
vwf
|
von Willebrand factor |
| chr5_+_52067723 | 0.05 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr8_-_48770163 | 0.05 |
ENSDART00000160959
|
pimr184
|
Pim proto-oncogene, serine/threonine kinase, related 184 |
| chr1_+_27070760 | 0.05 |
ENSDART00000186790
|
bnc2
|
basonuclin 2 |
| chr8_-_47818282 | 0.05 |
ENSDART00000137149
|
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr21_-_19919020 | 0.05 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr2_-_24068848 | 0.05 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr8_-_17926814 | 0.05 |
ENSDART00000147344
|
lhx8b
|
LIM homeobox 8b |
| chr4_-_44508975 | 0.04 |
ENSDART00000180528
|
si:dkeyp-100h4.1
|
si:dkeyp-100h4.1 |
| chr24_+_12200260 | 0.04 |
ENSDART00000181391
|
CR855257.2
|
|
| chr22_+_2598998 | 0.04 |
ENSDART00000176665
|
CU570894.1
|
|
| chr20_+_9211237 | 0.04 |
ENSDART00000139527
|
si:ch211-59d15.4
|
si:ch211-59d15.4 |
| chr4_-_71554068 | 0.04 |
ENSDART00000167414
|
si:dkey-27n6.1
|
si:dkey-27n6.1 |
| chr9_+_23511913 | 0.04 |
ENSDART00000137759
|
cntnap5a
|
contactin associated protein-like 5a |
| chr7_+_4583139 | 0.04 |
ENSDART00000143777
|
si:dkey-83f18.4
|
si:dkey-83f18.4 |
| chr1_-_9134045 | 0.03 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr23_+_20863145 | 0.03 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr12_+_44610912 | 0.03 |
ENSDART00000179030
ENSDART00000178008 |
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr11_+_6764207 | 0.03 |
ENSDART00000104292
|
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr8_-_17926620 | 0.03 |
ENSDART00000187864
|
lhx8b
|
LIM homeobox 8b |
| chr11_+_14295011 | 0.03 |
ENSDART00000060226
|
si:ch211-262i1.4
|
si:ch211-262i1.4 |
| chr21_-_41065369 | 0.03 |
ENSDART00000143749
|
larsb
|
leucyl-tRNA synthetase b |
| chr17_-_2386569 | 0.03 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr5_-_63286077 | 0.03 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr18_-_22717565 | 0.02 |
ENSDART00000142185
ENSDART00000059945 |
si:ch73-113g13.1
|
si:ch73-113g13.1 |
| chr21_-_5799122 | 0.02 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr6_+_27315173 | 0.02 |
ENSDART00000108831
|
espnla
|
espin like a |
| chr19_+_3215466 | 0.02 |
ENSDART00000181288
|
zgc:86598
|
zgc:86598 |
| chr16_+_38201840 | 0.02 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr18_+_2554666 | 0.02 |
ENSDART00000167218
|
p2ry2.1
|
purinergic receptor P2Y2, tandem duplicate 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tp53
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.2 | 1.7 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.2 | 0.8 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.2 | 0.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 0.9 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.2 | 1.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.7 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 1.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.5 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.2 | GO:0043525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.4 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.7 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.5 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.3 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.0 | 1.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.5 | GO:0043901 | negative regulation of immune effector process(GO:0002698) negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.2 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.5 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.7 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.4 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.5 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 1.0 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 0.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 1.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 1.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 0.8 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 0.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.5 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 1.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.2 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.3 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.6 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.7 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 1.0 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |