Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
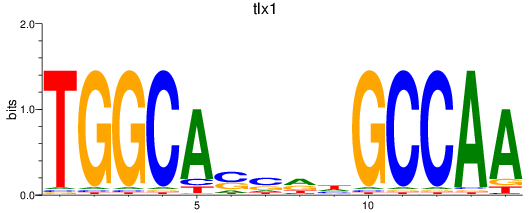
Results for tlx1
Z-value: 1.05
Transcription factors associated with tlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tlx1
|
ENSDARG00000003965 | T cell leukemia homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tlx1 | dr11_v1_chr13_-_28272299_28272299 | 0.83 | 5.9e-03 | Click! |
Activity profile of tlx1 motif
Sorted Z-values of tlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_30404096 | 2.36 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr2_+_55982940 | 2.22 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr19_-_30403922 | 1.92 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr20_+_10545514 | 1.90 |
ENSDART00000153667
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr20_+_10539293 | 1.90 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr17_+_52822422 | 1.78 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr9_-_22831836 | 1.77 |
ENSDART00000142585
|
neb
|
nebulin |
| chr20_+_16750177 | 1.76 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr22_+_22888 | 1.70 |
ENSDART00000082471
|
mfap2
|
microfibril associated protein 2 |
| chr12_+_27127139 | 1.61 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr1_-_58059134 | 1.54 |
ENSDART00000160970
|
caspb
|
caspase b |
| chr1_+_4101741 | 1.52 |
ENSDART00000163793
|
slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr16_+_23397785 | 1.47 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr1_+_49568335 | 1.42 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr7_-_35126374 | 1.40 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr17_+_52822831 | 1.38 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr7_+_14291323 | 1.38 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr19_+_9004256 | 1.31 |
ENSDART00000186401
ENSDART00000091443 |
si:ch211-81a5.1
|
si:ch211-81a5.1 |
| chr22_+_38229321 | 1.24 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr4_-_16406737 | 1.21 |
ENSDART00000013085
|
dcn
|
decorin |
| chr17_+_37645633 | 1.19 |
ENSDART00000085500
|
tmem229b
|
transmembrane protein 229B |
| chr22_+_11857356 | 1.18 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr2_-_8017579 | 1.16 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr9_-_34296406 | 1.15 |
ENSDART00000125451
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr20_+_34320635 | 1.14 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr1_-_22757145 | 1.13 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr3_-_5664123 | 1.10 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr13_-_9895564 | 1.06 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr6_-_13114406 | 1.05 |
ENSDART00000188015
|
zgc:194469
|
zgc:194469 |
| chr7_-_24472991 | 1.04 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr24_+_25692802 | 1.02 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr19_+_30388186 | 0.98 |
ENSDART00000103474
|
tspan13b
|
tetraspanin 13b |
| chr17_+_38262408 | 0.94 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr5_-_57655092 | 0.90 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr6_-_35401282 | 0.90 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr6_-_49673476 | 0.89 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr8_-_16712111 | 0.88 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr8_+_41647539 | 0.87 |
ENSDART00000136492
ENSDART00000138799 ENSDART00000134404 |
si:ch211-158d24.4
|
si:ch211-158d24.4 |
| chr3_+_57038033 | 0.82 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr6_+_53349966 | 0.82 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr7_+_57109214 | 0.79 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr23_-_45504991 | 0.78 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr10_-_17232372 | 0.78 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr17_+_52823015 | 0.75 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr4_+_74131530 | 0.74 |
ENSDART00000174125
|
CABZ01072523.1
|
|
| chr22_-_11438627 | 0.73 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr1_+_10297027 | 0.73 |
ENSDART00000152562
|
eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr2_-_17827983 | 0.73 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr21_-_34658266 | 0.70 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr9_-_24242592 | 0.68 |
ENSDART00000039399
|
cavin2a
|
caveolae associated protein 2a |
| chr6_-_34006917 | 0.67 |
ENSDART00000190702
ENSDART00000184003 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr1_-_22756898 | 0.64 |
ENSDART00000158915
|
prom1b
|
prominin 1 b |
| chr8_-_53108207 | 0.63 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr11_+_38332419 | 0.63 |
ENSDART00000005864
|
CDK18
|
si:dkey-166c18.1 |
| chr13_+_28761998 | 0.63 |
ENSDART00000146581
|
prom2
|
prominin 2 |
| chr1_-_28629471 | 0.63 |
ENSDART00000121758
|
ednrba
|
endothelin receptor Ba |
| chr23_-_20100436 | 0.62 |
ENSDART00000143445
|
si:dkey-32e6.6
|
si:dkey-32e6.6 |
| chr3_+_24482999 | 0.60 |
ENSDART00000059179
|
nptxra
|
neuronal pentraxin receptor a |
| chr15_-_34892664 | 0.54 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr11_+_37265692 | 0.51 |
ENSDART00000184691
|
il17rc
|
interleukin 17 receptor C |
| chr8_-_34052019 | 0.50 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr11_+_14284866 | 0.50 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr12_+_35654749 | 0.49 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr18_-_39473055 | 0.49 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr17_-_22067451 | 0.49 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr16_+_14029283 | 0.49 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr13_-_31829786 | 0.49 |
ENSDART00000138667
|
sertad4
|
SERTA domain containing 4 |
| chr2_-_21618629 | 0.48 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr13_-_32577386 | 0.47 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr7_-_22132265 | 0.46 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr11_-_34147205 | 0.43 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr9_-_25089536 | 0.42 |
ENSDART00000143479
|
rubcnl
|
RUN and cysteine rich domain containing beclin 1 interacting protein like |
| chr5_-_16218777 | 0.42 |
ENSDART00000141698
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr9_-_23358597 | 0.42 |
ENSDART00000046423
|
arl4cb
|
ADP-ribosylation factor-like 4Cb |
| chr1_-_56363108 | 0.41 |
ENSDART00000021878
|
gcgrb
|
glucagon receptor b |
| chr9_-_44939104 | 0.40 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr4_+_61171310 | 0.38 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr7_-_52096498 | 0.38 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr20_-_26066020 | 0.38 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr20_+_54383838 | 0.36 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr13_+_23162447 | 0.36 |
ENSDART00000180209
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr6_-_21873266 | 0.36 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr13_+_37040789 | 0.36 |
ENSDART00000063412
|
esr2b
|
estrogen receptor 2b |
| chr4_-_64709908 | 0.35 |
ENSDART00000161032
|
si:dkey-9i5.2
|
si:dkey-9i5.2 |
| chr24_-_21819010 | 0.35 |
ENSDART00000091096
|
CR352265.1
|
|
| chr6_+_39184236 | 0.34 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr4_-_38033800 | 0.33 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr17_-_45040813 | 0.33 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr24_+_7884880 | 0.33 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr7_-_41468751 | 0.33 |
ENSDART00000150146
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr6_+_28141135 | 0.32 |
ENSDART00000113405
ENSDART00000179795 |
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr2_-_59303338 | 0.31 |
ENSDART00000100987
ENSDART00000140840 |
ftr35
|
finTRIM family, member 35 |
| chr15_-_14469704 | 0.31 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr10_+_7636811 | 0.29 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr9_+_16449398 | 0.28 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr21_-_2707768 | 0.27 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr2_-_59285085 | 0.27 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr10_+_8091144 | 0.25 |
ENSDART00000143848
ENSDART00000075485 |
sub1a
|
SUB1 homolog, transcriptional regulator a |
| chr17_-_22048233 | 0.24 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
| chr2_-_53925308 | 0.23 |
ENSDART00000190705
|
FO834855.1
|
|
| chr11_+_24251141 | 0.23 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr6_+_2951645 | 0.23 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr2_-_59285407 | 0.22 |
ENSDART00000181616
|
ftr34
|
finTRIM family, member 34 |
| chr20_-_33976305 | 0.19 |
ENSDART00000111022
|
sele
|
selectin E |
| chr16_+_16849220 | 0.18 |
ENSDART00000047409
ENSDART00000142155 |
myh14
|
myosin, heavy chain 14, non-muscle |
| chr7_+_52135791 | 0.17 |
ENSDART00000098705
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr8_+_53423408 | 0.17 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr18_-_2433011 | 0.16 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr15_-_18091157 | 0.15 |
ENSDART00000153848
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr20_-_20410029 | 0.15 |
ENSDART00000192177
ENSDART00000063483 |
prkchb
|
protein kinase C, eta, b |
| chr4_-_58448785 | 0.14 |
ENSDART00000172359
|
si:ch211-212k5.3
|
si:ch211-212k5.3 |
| chr12_-_37941733 | 0.14 |
ENSDART00000130167
|
CABZ01046949.1
|
|
| chr6_+_41957280 | 0.12 |
ENSDART00000050114
|
oxtr
|
oxytocin receptor |
| chr5_+_35502290 | 0.11 |
ENSDART00000191222
|
CR762390.1
|
|
| chr13_-_41482064 | 0.11 |
ENSDART00000188322
ENSDART00000164732 |
pcdh15a
|
protocadherin-related 15a |
| chr6_+_28140816 | 0.11 |
ENSDART00000137907
|
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr3_-_37681824 | 0.11 |
ENSDART00000185858
|
gpatch8
|
G patch domain containing 8 |
| chr6_+_26346686 | 0.10 |
ENSDART00000154183
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr20_+_48739154 | 0.10 |
ENSDART00000100262
|
zgc:110348
|
zgc:110348 |
| chr3_+_33918510 | 0.09 |
ENSDART00000055248
|
alkbh7
|
alkB homolog 7 |
| chr20_-_2134620 | 0.07 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr3_-_40528333 | 0.07 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr4_-_51460013 | 0.07 |
ENSDART00000193382
|
CR628395.1
|
|
| chr7_+_57108823 | 0.07 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr17_-_31308658 | 0.06 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr7_-_51953807 | 0.06 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr15_-_38202630 | 0.05 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr5_-_57526807 | 0.04 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr3_+_61391636 | 0.04 |
ENSDART00000126417
|
bri3
|
brain protein I3 |
| chr10_-_15672862 | 0.01 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr23_-_18572685 | 0.01 |
ENSDART00000047175
|
ssr4
|
signal sequence receptor, delta |
| chr17_-_7496341 | 0.01 |
ENSDART00000186907
|
si:ch211-278p9.3
|
si:ch211-278p9.3 |
| chr19_+_22727940 | 0.00 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
| chr12_-_36521947 | 0.00 |
ENSDART00000152946
|
unc13d
|
unc-13 homolog D (C. elegans) |
Network of associatons between targets according to the STRING database.
First level regulatory network of tlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.3 | 1.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 0.9 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.2 | 1.1 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 1.4 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.2 | 1.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 1.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.4 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.4 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.1 | 0.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.4 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 5.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.5 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 1.5 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:0021985 | parturition(GO:0007567) neurohypophysis development(GO:0021985) multi-multicellular organism process(GO:0044706) maternal process involved in parturition(GO:0060137) |
| 0.0 | 3.9 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.3 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 1.2 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 0.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0055062 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.0 | 0.9 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 1.7 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.0 | 1.2 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.7 | GO:0072175 | epithelial tube formation(GO:0072175) |
| 0.0 | 0.1 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.0 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.7 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 1.5 | GO:0008544 | epidermis development(GO:0008544) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.3 | 2.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 0.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.1 | GO:0044420 | extracellular matrix component(GO:0044420) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 0.9 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.2 | 1.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 1.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.4 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 0.4 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 1.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 2.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.7 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 4.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 2.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |