Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
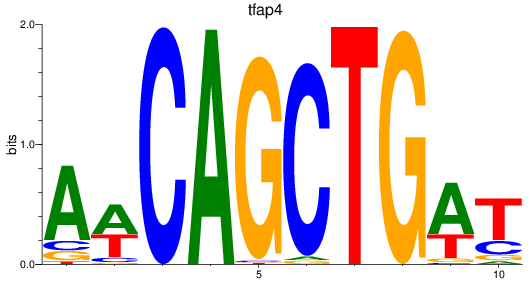
Results for tfap4
Z-value: 2.19
Transcription factors associated with tfap4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap4
|
ENSDARG00000103923 | transcription factor AP-4 (activating enhancer binding protein 4) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap4 | dr11_v1_chr3_-_12227359_12227359 | -0.18 | 6.4e-01 | Click! |
Activity profile of tfap4 motif
Sorted Z-values of tfap4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_20693743 | 5.79 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr7_+_17947217 | 5.49 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr9_-_5351017 | 4.74 |
ENSDART00000082260
|
abhd13
|
abhydrolase domain containing 13 |
| chr15_+_38299563 | 4.30 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr11_+_45448212 | 4.06 |
ENSDART00000173341
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr4_-_17278904 | 3.60 |
ENSDART00000178686
ENSDART00000135730 |
lrmp
|
lymphoid-restricted membrane protein |
| chr15_+_38308421 | 3.04 |
ENSDART00000129941
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr13_-_2336584 | 2.93 |
ENSDART00000113692
|
tceb3l
|
transcription elongation factor B (SIII), polypeptide 3, like |
| chr14_-_34605607 | 2.66 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr5_-_9216758 | 2.57 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr12_-_10505986 | 2.57 |
ENSDART00000152672
|
zgc:152977
|
zgc:152977 |
| chr10_+_399363 | 2.52 |
ENSDART00000147449
|
si:ch211-242f23.8
|
si:ch211-242f23.8 |
| chr14_-_34605804 | 2.46 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr12_+_4920451 | 2.38 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr10_-_25217347 | 2.36 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr14_+_35464994 | 2.35 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr17_-_17759138 | 2.31 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr21_-_32060993 | 2.17 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr17_+_43868441 | 2.09 |
ENSDART00000134272
|
zgc:66313
|
zgc:66313 |
| chr17_-_38887424 | 2.07 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr5_+_37903790 | 2.07 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr12_-_33359654 | 1.99 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_-_55779927 | 1.99 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr19_+_10661520 | 1.97 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr16_-_21489514 | 1.94 |
ENSDART00000149525
ENSDART00000148517 ENSDART00000146914 ENSDART00000186493 ENSDART00000193081 ENSDART00000186017 |
mpp6a
|
membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) |
| chr5_+_65970235 | 1.94 |
ENSDART00000166432
|
slc2a8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr10_+_31809226 | 1.93 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr14_+_50918769 | 1.92 |
ENSDART00000146918
|
rnf44
|
ring finger protein 44 |
| chr13_+_40815012 | 1.91 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr21_+_4204860 | 1.91 |
ENSDART00000146541
|
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr7_+_73308566 | 1.90 |
ENSDART00000187039
ENSDART00000174244 |
CABZ01081777.1
|
|
| chr9_-_1200187 | 1.90 |
ENSDART00000158760
|
ino80da
|
INO80 complex subunit Da |
| chr23_-_18057851 | 1.90 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr16_-_24642814 | 1.88 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr19_+_14454306 | 1.86 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr3_+_14571813 | 1.86 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr17_+_14965570 | 1.84 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr9_+_38168012 | 1.84 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr2_+_37245382 | 1.82 |
ENSDART00000004626
|
sec62
|
SEC62 homolog, preprotein translocation factor |
| chr7_-_69795488 | 1.81 |
ENSDART00000162414
|
USP53 (1 of many)
|
ubiquitin specific peptidase 53 |
| chr9_-_27717006 | 1.81 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr6_-_9952103 | 1.81 |
ENSDART00000065475
|
zp2l2
|
zona pellucida glycoprotein 2, like 2 |
| chr6_+_45494227 | 1.79 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| chr8_-_22326073 | 1.79 |
ENSDART00000084965
|
cep104
|
centrosomal protein 104 |
| chr5_-_11809404 | 1.79 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr5_-_68333081 | 1.78 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr21_-_3606539 | 1.77 |
ENSDART00000062418
|
dym
|
dymeclin |
| chr2_+_24700922 | 1.75 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr2_+_44518636 | 1.75 |
ENSDART00000153733
|
pask
|
PAS domain containing serine/threonine kinase |
| chr5_+_31779911 | 1.74 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr18_-_399554 | 1.73 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr8_+_48966165 | 1.73 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr14_+_16083818 | 1.72 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr8_+_48965767 | 1.72 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr16_+_41067586 | 1.71 |
ENSDART00000181876
|
scap
|
SREBF chaperone |
| chr12_-_3840664 | 1.67 |
ENSDART00000160967
|
taok2b
|
TAO kinase 2b |
| chr13_+_28580357 | 1.66 |
ENSDART00000007211
|
wbp1la
|
WW domain binding protein 1-like a |
| chr15_-_5624361 | 1.66 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr19_-_2085027 | 1.66 |
ENSDART00000063615
|
snx13
|
sorting nexin 13 |
| chr5_-_29195063 | 1.65 |
ENSDART00000109926
|
man1b1b
|
mannosidase, alpha, class 1B, member 1b |
| chr21_-_21515466 | 1.65 |
ENSDART00000147593
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr21_+_76739 | 1.65 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr6_-_8360918 | 1.64 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr16_-_30655980 | 1.64 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr11_-_11792766 | 1.63 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr15_-_31147301 | 1.62 |
ENSDART00000157145
ENSDART00000155473 ENSDART00000048103 |
ksr1b
|
kinase suppressor of ras 1b |
| chr3_-_40836081 | 1.62 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr5_-_65121747 | 1.61 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr23_-_3511630 | 1.59 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr2_-_17114852 | 1.58 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr22_-_16443199 | 1.58 |
ENSDART00000006290
ENSDART00000193335 |
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr19_-_4793263 | 1.57 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr5_-_32336613 | 1.56 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_-_16177603 | 1.56 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr23_+_25893020 | 1.56 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr16_-_42750295 | 1.56 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr9_-_39624173 | 1.55 |
ENSDART00000180106
ENSDART00000126766 |
erbb4b
|
erb-b2 receptor tyrosine kinase 4b |
| chr9_+_46745348 | 1.55 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr7_-_59515569 | 1.55 |
ENSDART00000163343
ENSDART00000165457 ENSDART00000163745 |
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr13_-_17464362 | 1.55 |
ENSDART00000145499
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr22_-_3299100 | 1.54 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr16_-_22544047 | 1.53 |
ENSDART00000131657
|
cgna
|
cingulin a |
| chr17_-_41798856 | 1.51 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr3_+_7771420 | 1.51 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr23_-_19831739 | 1.51 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr20_-_35508805 | 1.49 |
ENSDART00000169538
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr20_+_20731052 | 1.48 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr3_-_21118969 | 1.48 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr9_+_21306902 | 1.48 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr1_+_35473219 | 1.47 |
ENSDART00000109678
ENSDART00000181635 |
usp38
|
ubiquitin specific peptidase 38 |
| chr6_+_59991076 | 1.47 |
ENSDART00000163575
|
CABZ01100888.1
|
|
| chr7_+_34794829 | 1.44 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr8_+_25145464 | 1.44 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr10_+_15340768 | 1.43 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr16_-_4610255 | 1.43 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr20_-_40367493 | 1.43 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr9_+_33154841 | 1.42 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr8_-_4010887 | 1.41 |
ENSDART00000163678
|
mtmr3
|
myotubularin related protein 3 |
| chr13_+_16521898 | 1.40 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr10_-_10969596 | 1.39 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr11_-_44999858 | 1.39 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr19_-_20446756 | 1.38 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr1_+_35473397 | 1.38 |
ENSDART00000158799
|
usp38
|
ubiquitin specific peptidase 38 |
| chr12_-_33579873 | 1.38 |
ENSDART00000184661
|
tdrkh
|
tudor and KH domain containing |
| chr2_-_20866758 | 1.37 |
ENSDART00000165374
|
tpra
|
translocated promoter region a, nuclear basket protein |
| chr15_-_9421481 | 1.37 |
ENSDART00000189045
ENSDART00000177158 |
sacs
|
sacsin molecular chaperone |
| chr5_-_16405651 | 1.36 |
ENSDART00000163942
|
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr10_-_10969444 | 1.35 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr1_+_41666611 | 1.35 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr24_+_5811808 | 1.34 |
ENSDART00000132428
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr5_-_11809710 | 1.34 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr8_+_23382568 | 1.33 |
ENSDART00000129167
|
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr2_+_20866898 | 1.33 |
ENSDART00000150086
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr10_-_35257458 | 1.33 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr23_-_18057270 | 1.32 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr10_-_11840353 | 1.31 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr10_+_1849874 | 1.31 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr13_-_17464654 | 1.31 |
ENSDART00000140312
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr4_+_5832311 | 1.30 |
ENSDART00000121743
ENSDART00000158233 ENSDART00000165187 |
si:ch73-352p4.5
|
si:ch73-352p4.5 |
| chr12_+_33894396 | 1.29 |
ENSDART00000130853
ENSDART00000152988 |
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr14_-_16754262 | 1.28 |
ENSDART00000001159
|
mgat4b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr2_-_17115256 | 1.27 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr3_-_34753605 | 1.26 |
ENSDART00000000160
|
thraa
|
thyroid hormone receptor alpha a |
| chr12_+_31638045 | 1.25 |
ENSDART00000184216
ENSDART00000183645 ENSDART00000153129 |
dnmbp
|
dynamin binding protein |
| chr13_-_33114933 | 1.25 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr11_+_24314148 | 1.24 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr9_+_21795917 | 1.24 |
ENSDART00000169069
|
rev1
|
REV1, polymerase (DNA directed) |
| chr23_+_27778670 | 1.23 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr23_+_44461493 | 1.23 |
ENSDART00000149854
|
si:ch1073-228j22.1
|
si:ch1073-228j22.1 |
| chr5_-_37341044 | 1.23 |
ENSDART00000084675
|
wdr44
|
WD repeat domain 44 |
| chr23_-_18057553 | 1.22 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr12_+_10116912 | 1.22 |
ENSDART00000189630
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr5_-_2636078 | 1.21 |
ENSDART00000122274
|
cita
|
citron rho-interacting serine/threonine kinase a |
| chr17_+_51940768 | 1.21 |
ENSDART00000053422
|
ttll5
|
tubulin tyrosine ligase-like family, member 5 |
| chr3_-_33113879 | 1.20 |
ENSDART00000044677
|
rarab
|
retinoic acid receptor, alpha b |
| chr17_-_18898115 | 1.20 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr3_+_41731527 | 1.20 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr23_-_21748805 | 1.20 |
ENSDART00000148128
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr6_+_33076839 | 1.19 |
ENSDART00000073755
ENSDART00000122242 |
pomgnt1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr8_-_14609284 | 1.19 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr24_-_27256338 | 1.19 |
ENSDART00000105768
|
zmynd11
|
zinc finger, MYND-type containing 11 |
| chr16_-_36196700 | 1.18 |
ENSDART00000172324
|
capn7
|
calpain 7 |
| chr16_-_54971277 | 1.18 |
ENSDART00000113358
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr2_-_11027258 | 1.18 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr19_+_43684376 | 1.18 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr19_-_42503143 | 1.18 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr16_-_21903083 | 1.16 |
ENSDART00000165849
|
setdb1b
|
SET domain, bifurcated 1b |
| chr12_-_25150239 | 1.16 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr24_+_5811350 | 1.16 |
ENSDART00000178238
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr9_+_2522797 | 1.16 |
ENSDART00000186786
ENSDART00000147034 |
gpr155a
|
G protein-coupled receptor 155a |
| chr15_-_30857350 | 1.16 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr18_-_7948188 | 1.15 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr2_-_22286828 | 1.15 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr20_-_28361574 | 1.15 |
ENSDART00000103352
|
vps18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr10_+_44373349 | 1.14 |
ENSDART00000172191
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr1_-_14258409 | 1.14 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr8_-_410199 | 1.13 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr5_-_24245218 | 1.13 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr11_-_3533356 | 1.13 |
ENSDART00000161972
|
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr6_-_7776612 | 1.11 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr24_+_35183595 | 1.11 |
ENSDART00000075142
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr18_-_42071876 | 1.11 |
ENSDART00000193288
ENSDART00000189021 ENSDART00000019245 |
arhgap42b
|
Rho GTPase activating protein 42b |
| chr19_-_10214264 | 1.10 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr18_-_41232297 | 1.10 |
ENSDART00000036928
|
fbxo36a
|
F-box protein 36a |
| chr19_+_15509372 | 1.10 |
ENSDART00000151530
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr15_+_24676905 | 1.09 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr16_+_27383717 | 1.09 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr2_-_37098785 | 1.09 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr5_+_30392148 | 1.09 |
ENSDART00000086765
|
stk36
|
serine/threonine kinase 36 (fused homolog, Drosophila) |
| chr19_+_4968947 | 1.09 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr16_+_23796612 | 1.08 |
ENSDART00000131698
|
rab13
|
RAB13, member RAS oncogene family |
| chr18_+_6641542 | 1.07 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr9_+_2020667 | 1.07 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr18_-_19819812 | 1.07 |
ENSDART00000060344
|
aagab
|
alpha and gamma adaptin binding protein |
| chr18_+_45526585 | 1.07 |
ENSDART00000138511
|
kifc3
|
kinesin family member C3 |
| chr15_+_40188076 | 1.06 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr10_+_37182626 | 1.05 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr17_-_8570257 | 1.05 |
ENSDART00000154713
ENSDART00000121488 |
fzd3b
|
frizzled class receptor 3b |
| chr24_+_14937205 | 1.05 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr2_+_25560556 | 1.05 |
ENSDART00000133623
|
pld1a
|
phospholipase D1a |
| chr23_+_13528550 | 1.05 |
ENSDART00000099903
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr6_+_18520859 | 1.05 |
ENSDART00000158263
|
si:dkey-10p5.10
|
si:dkey-10p5.10 |
| chr23_-_21758253 | 1.05 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr7_+_66634167 | 1.05 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr24_+_18689980 | 1.04 |
ENSDART00000160941
ENSDART00000180303 ENSDART00000190435 |
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr9_+_50110763 | 1.04 |
ENSDART00000162990
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr15_+_47746176 | 1.03 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr24_-_31123365 | 1.03 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr24_-_27256673 | 1.03 |
ENSDART00000181182
|
zmynd11
|
zinc finger, MYND-type containing 11 |
| chr20_+_43691208 | 1.02 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr19_-_15192840 | 1.00 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr1_+_57348756 | 1.00 |
ENSDART00000063750
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr7_+_41265618 | 1.00 |
ENSDART00000173688
|
scrib
|
scribbled planar cell polarity protein |
| chr6_-_54348568 | 1.00 |
ENSDART00000156501
|
zgc:101577
|
zgc:101577 |
| chr20_+_715739 | 1.00 |
ENSDART00000136768
|
myo6a
|
myosin VIa |
| chr17_+_24446705 | 1.00 |
ENSDART00000163221
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr1_+_35956435 | 0.98 |
ENSDART00000085021
ENSDART00000148505 |
mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr19_-_23249822 | 0.98 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr2_-_31800521 | 0.98 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr1_-_50293946 | 0.98 |
ENSDART00000053028
ENSDART00000125099 |
tbck
|
TBC1 domain containing kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.6 | 1.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.6 | 1.8 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.6 | 2.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.6 | 1.7 | GO:0010656 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.5 | 2.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.5 | 1.4 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.4 | 2.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.6 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.4 | 2.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 1.3 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.3 | 1.0 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.3 | 1.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.3 | 1.6 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.3 | 0.9 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 1.2 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.3 | 1.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.3 | 1.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 1.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.3 | 1.1 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.3 | 1.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.3 | 1.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 2.0 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.2 | 0.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 1.0 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 2.9 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 1.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 1.2 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.2 | 1.1 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.2 | 1.1 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.2 | 2.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 1.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 3.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 0.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 2.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 1.4 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.2 | 1.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 0.6 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 1.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.2 | 0.7 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 1.8 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.2 | 0.9 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 1.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 0.8 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.2 | 2.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 1.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 1.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 0.9 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 0.8 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.2 | 0.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.8 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.2 | 1.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 1.9 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.6 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 1.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.2 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.8 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.1 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.8 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 0.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 1.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.8 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.0 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 1.0 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 1.0 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.9 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.8 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.8 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.8 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.1 | 0.9 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 1.0 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.2 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 1.0 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.6 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.1 | 0.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.1 | 0.2 | GO:1903646 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 0.8 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 2.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.0 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 1.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 1.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.9 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 0.6 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.0 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.9 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 2.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 2.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 1.4 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.0 | 0.2 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0015824 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.0 | 0.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 2.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 1.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.9 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 2.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 2.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.1 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.7 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.3 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 2.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 8.4 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 1.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 3.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.7 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 2.3 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 3.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.8 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0015810 | C4-dicarboxylate transport(GO:0015740) acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.9 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.9 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 1.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.6 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 3.6 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 1.1 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.7 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.5 | 1.4 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.4 | 5.1 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 1.8 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.4 | 1.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.3 | 1.4 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.3 | 3.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 0.8 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 1.5 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 2.0 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 0.7 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.2 | 0.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 0.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 0.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 0.5 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.2 | 1.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 1.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 0.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.8 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 2.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 1.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.2 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 6.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 2.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 2.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.8 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 3.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 3.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 2.3 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.2 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 2.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 15.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.7 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 2.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.4 | GO:0005764 | lysosome(GO:0005764) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.5 | 2.5 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.5 | 1.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.4 | 1.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 1.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.4 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.3 | 1.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 1.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 1.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.3 | 3.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 1.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 1.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 0.9 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 2.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 4.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.9 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 0.9 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 2.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.5 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 1.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 2.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 1.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.7 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.2 | 0.7 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.7 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.2 | 1.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 0.9 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.2 | 1.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 5.8 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 1.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.5 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.5 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.4 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 1.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.5 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 6.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.6 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 2.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 4.2 | GO:0032934 | sterol binding(GO:0032934) |
| 0.1 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.9 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.5 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 2.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 3.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 4.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.7 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.3 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.8 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 1.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 5.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.7 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 1.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 1.2 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.9 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.9 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.3 | 1.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 1.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 1.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 1.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 2.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |