Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
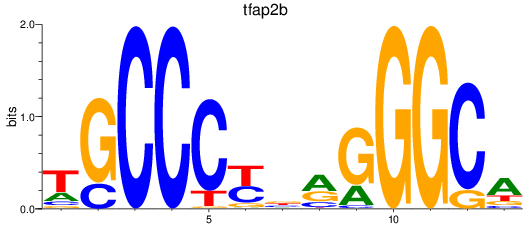
Results for tfap2b
Z-value: 1.06
Transcription factors associated with tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2b
|
ENSDARG00000012667 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2b | dr11_v1_chr20_-_47704973_47704973 | -0.50 | 1.7e-01 | Click! |
Activity profile of tfap2b motif
Sorted Z-values of tfap2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_33857911 | 1.14 |
ENSDART00000165370
|
nxph2b
|
neurexophilin 2b |
| chr4_-_77551860 | 1.08 |
ENSDART00000188176
|
AL935186.6
|
|
| chr4_-_77563411 | 1.02 |
ENSDART00000186841
|
AL935186.8
|
|
| chr7_-_7420301 | 1.01 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr4_-_20115570 | 0.97 |
ENSDART00000134528
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr15_-_32131 | 0.83 |
ENSDART00000099074
ENSDART00000164323 |
CYP2C9
|
si:zfos-411a11.2 |
| chr17_-_26867725 | 0.82 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr12_+_30360184 | 0.81 |
ENSDART00000190718
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr13_+_32148338 | 0.74 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr17_-_26868169 | 0.73 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr12_-_8958353 | 0.62 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr7_+_48667081 | 0.60 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr12_+_30360579 | 0.57 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr18_+_24919614 | 0.52 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr18_+_40364732 | 0.52 |
ENSDART00000123661
ENSDART00000130847 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr15_-_20710 | 0.51 |
ENSDART00000161218
|
cyp2y3
|
cytochrome P450, family 2, subfamily Y, polypeptide 3 |
| chr12_+_5708400 | 0.48 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr9_-_15424639 | 0.48 |
ENSDART00000124346
|
fn1a
|
fibronectin 1a |
| chr7_-_38570299 | 0.45 |
ENSDART00000143815
|
celf1
|
cugbp, Elav-like family member 1 |
| chr16_+_26612401 | 0.41 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr7_-_38570878 | 0.40 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr10_-_5135788 | 0.39 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr3_-_506241 | 0.38 |
ENSDART00000150141
|
si:dkey-12f6.5
|
si:dkey-12f6.5 |
| chr22_+_508290 | 0.37 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr8_+_23861461 | 0.37 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr25_+_17871089 | 0.36 |
ENSDART00000133725
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr12_+_638435 | 0.36 |
ENSDART00000152508
|
si:ch211-176g6.1
|
si:ch211-176g6.1 |
| chr23_-_1017428 | 0.35 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr1_-_55248496 | 0.32 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr8_+_24281512 | 0.31 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr23_-_1017605 | 0.28 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr4_+_58667348 | 0.26 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr21_-_27362938 | 0.26 |
ENSDART00000131297
|
rin1a
|
Ras and Rab interactor 1a |
| chr7_+_529522 | 0.24 |
ENSDART00000190811
|
nrxn2b
|
neurexin 2b |
| chr7_+_34794829 | 0.23 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr21_+_21205667 | 0.22 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr25_-_20870900 | 0.21 |
ENSDART00000171288
|
magi2b
|
membrane associated guanylate kinase, WW and PDZ domain containing 2b |
| chr5_-_15948833 | 0.21 |
ENSDART00000051649
ENSDART00000124467 |
xbp1
|
X-box binding protein 1 |
| chr1_+_5275811 | 0.21 |
ENSDART00000189676
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr9_+_22764235 | 0.21 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr24_+_16149251 | 0.20 |
ENSDART00000188289
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr17_-_19019635 | 0.20 |
ENSDART00000126666
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr21_-_34926619 | 0.20 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr22_-_9906790 | 0.19 |
ENSDART00000105936
|
si:dkey-253d23.8
|
si:dkey-253d23.8 |
| chr1_-_43987873 | 0.19 |
ENSDART00000108821
|
CR385050.1
|
|
| chr24_+_42074143 | 0.18 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr4_+_5835393 | 0.18 |
ENSDART00000055414
|
pex26
|
peroxisomal biogenesis factor 26 |
| chr6_-_59942335 | 0.18 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr14_-_51039702 | 0.16 |
ENSDART00000178932
|
cltb
|
clathrin, light chain B |
| chr25_+_30460378 | 0.16 |
ENSDART00000016310
|
banp
|
BTG3 associated nuclear protein |
| chr3_-_43646733 | 0.16 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr25_+_6471401 | 0.14 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr10_+_6383270 | 0.13 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr1_-_49544601 | 0.13 |
ENSDART00000145960
|
slc47a2.2
|
solute carrier family 47 (multidrug and toxin extrusion), member 2.2 |
| chr16_+_20496691 | 0.12 |
ENSDART00000182737
ENSDART00000078984 |
cpvl
|
carboxypeptidase, vitellogenic-like |
| chr6_+_9867426 | 0.12 |
ENSDART00000151749
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr2_+_9918678 | 0.12 |
ENSDART00000081253
|
acbd5b
|
acyl-CoA binding domain containing 5b |
| chr2_+_9918935 | 0.12 |
ENSDART00000140434
|
acbd5b
|
acyl-CoA binding domain containing 5b |
| chr14_+_31509922 | 0.11 |
ENSDART00000124499
|
hprt1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr6_+_38381957 | 0.10 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr16_-_20435475 | 0.10 |
ENSDART00000139776
|
chn2
|
chimerin 2 |
| chr7_+_21887787 | 0.10 |
ENSDART00000162252
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr25_-_32363341 | 0.09 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr4_+_54439143 | 0.09 |
ENSDART00000170168
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr4_-_63980140 | 0.09 |
ENSDART00000160818
|
znf1091
|
zinc finger protein 1091 |
| chr16_+_54209504 | 0.08 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr12_-_32013125 | 0.08 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr6_+_52931841 | 0.07 |
ENSDART00000174358
|
si:dkeyp-3f10.12
|
si:dkeyp-3f10.12 |
| chr15_-_3736773 | 0.07 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr7_-_48667056 | 0.07 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr20_-_47188966 | 0.07 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr6_-_2171818 | 0.06 |
ENSDART00000181082
ENSDART00000187556 |
PXMP4
|
wu:fe02h09 |
| chr9_-_48864942 | 0.06 |
ENSDART00000113855
ENSDART00000189064 |
nostrin
|
nitric oxide synthase trafficking |
| chr17_-_29771639 | 0.05 |
ENSDART00000086201
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr1_-_23293261 | 0.05 |
ENSDART00000122648
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr16_-_49846359 | 0.05 |
ENSDART00000184246
|
znf385d
|
zinc finger protein 385D |
| chr20_+_51199666 | 0.05 |
ENSDART00000169321
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr20_+_30578967 | 0.05 |
ENSDART00000010494
|
fgfr1op
|
FGFR1 oncogene partner |
| chr8_-_37056622 | 0.05 |
ENSDART00000111513
|
si:ch211-218o21.4
|
si:ch211-218o21.4 |
| chr4_+_37992753 | 0.04 |
ENSDART00000193890
|
CR759843.2
|
|
| chr23_-_27701361 | 0.04 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr1_+_32528097 | 0.04 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr11_-_35975026 | 0.03 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr4_-_60790123 | 0.02 |
ENSDART00000137702
|
si:dkey-254e13.6
|
si:dkey-254e13.6 |
| chr17_-_52609728 | 0.02 |
ENSDART00000103572
|
lrrc74a
|
leucine rich repeat containing 74A |
| chr2_-_9544161 | 0.02 |
ENSDART00000124425
|
slc25a24l
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24, like |
| chr9_+_33334501 | 0.02 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr2_+_39108339 | 0.02 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr5_+_45921234 | 0.00 |
ENSDART00000134355
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr24_+_37723362 | 0.00 |
ENSDART00000136836
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.2 | 0.9 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.5 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.6 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.6 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.3 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.5 | GO:0006953 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) acute-phase response(GO:0006953) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.2 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 1.0 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.2 | GO:0030910 | olfactory placode formation(GO:0030910) |
| 0.0 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 1.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.0 | GO:0071634 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.2 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.2 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.6 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |