Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
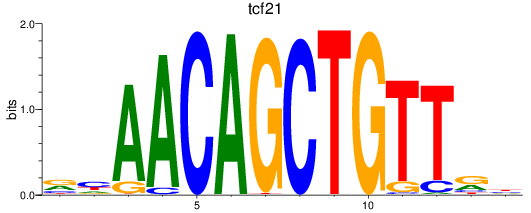
Results for tcf21
Z-value: 0.39
Transcription factors associated with tcf21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf21
|
ENSDARG00000036869 | transcription factor 21 |
|
tcf21
|
ENSDARG00000111209 | transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf21 | dr11_v1_chr23_-_31555696_31555696 | 0.05 | 8.9e-01 | Click! |
Activity profile of tcf21 motif
Sorted Z-values of tcf21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_32166702 | 0.35 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr9_-_35633827 | 0.27 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr16_+_23972126 | 0.20 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr13_+_4422795 | 0.18 |
ENSDART00000138211
|
si:ch211-130h14.4
|
si:ch211-130h14.4 |
| chr14_+_12109535 | 0.18 |
ENSDART00000054620
ENSDART00000121913 |
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr2_+_4208323 | 0.18 |
ENSDART00000167906
|
gata6
|
GATA binding protein 6 |
| chr5_+_4366431 | 0.17 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr11_-_35171162 | 0.17 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr19_-_19505167 | 0.17 |
ENSDART00000160582
|
creb5a
|
cAMP responsive element binding protein 5a |
| chr7_-_62244744 | 0.16 |
ENSDART00000192522
ENSDART00000170635 ENSDART00000073872 |
CCKAR
|
cholecystokinin A receptor |
| chr15_+_20403084 | 0.15 |
ENSDART00000141388
ENSDART00000152734 |
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr21_+_25932107 | 0.15 |
ENSDART00000185162
|
caln2
|
calneuron 2 |
| chr8_-_29706882 | 0.15 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr8_+_39724138 | 0.15 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr23_-_18057553 | 0.14 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr7_+_34231782 | 0.14 |
ENSDART00000173547
|
lctla
|
lactase-like a |
| chr9_+_32301456 | 0.13 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr13_+_24396666 | 0.13 |
ENSDART00000139197
ENSDART00000101200 |
zgc:153169
|
zgc:153169 |
| chr6_+_25261297 | 0.13 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr5_+_28849155 | 0.13 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr25_+_36045072 | 0.13 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr23_-_18057851 | 0.12 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr17_+_8323348 | 0.12 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr6_-_47838481 | 0.12 |
ENSDART00000185284
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr10_-_25217347 | 0.12 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr23_-_18057270 | 0.12 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr15_+_21254800 | 0.11 |
ENSDART00000142902
|
usf1
|
upstream transcription factor 1 |
| chr24_-_35707552 | 0.11 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr12_-_27242498 | 0.11 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr14_+_12110020 | 0.11 |
ENSDART00000192462
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr5_+_28858345 | 0.11 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr5_+_28830643 | 0.10 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr10_+_35491216 | 0.10 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr1_-_39895859 | 0.10 |
ENSDART00000135791
ENSDART00000035739 |
tmem134
|
transmembrane protein 134 |
| chr12_+_22404108 | 0.10 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr14_+_146857 | 0.09 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr11_-_5865744 | 0.09 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr11_-_35171768 | 0.09 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr9_+_13714379 | 0.09 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr8_-_1838315 | 0.09 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr19_+_35002479 | 0.09 |
ENSDART00000103266
|
wdyhv1
|
WDYHV motif containing 1 |
| chr12_+_20693743 | 0.09 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr5_+_25762271 | 0.09 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr5_+_63322093 | 0.08 |
ENSDART00000158086
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr9_-_44983666 | 0.08 |
ENSDART00000149704
|
itgb2
|
integrin, beta 2 |
| chr13_-_41155472 | 0.08 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr3_-_52661242 | 0.08 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr15_-_945804 | 0.08 |
ENSDART00000063257
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr7_+_30051488 | 0.08 |
ENSDART00000169410
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr15_+_11683114 | 0.08 |
ENSDART00000168233
|
si:dkey-31c13.1
|
si:dkey-31c13.1 |
| chr4_+_9612574 | 0.08 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr5_+_63305357 | 0.08 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr2_+_243778 | 0.08 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr6_+_23931236 | 0.08 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr21_-_41617372 | 0.08 |
ENSDART00000187171
|
BX005397.1
|
|
| chr1_-_52498146 | 0.08 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_+_10610791 | 0.08 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr5_-_23729590 | 0.08 |
ENSDART00000014831
|
si:dkey-110k5.10
|
si:dkey-110k5.10 |
| chr2_-_17114852 | 0.08 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr14_+_32837914 | 0.08 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr23_-_16692312 | 0.07 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr3_+_24537023 | 0.07 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr23_+_29908348 | 0.07 |
ENSDART00000186622
ENSDART00000150174 |
si:ch73-236e11.2
|
si:ch73-236e11.2 |
| chr8_+_40080728 | 0.07 |
ENSDART00000111506
|
lrrc75ba
|
leucine rich repeat containing 75Ba |
| chr8_-_25247284 | 0.07 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr10_+_10738880 | 0.07 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr16_+_41015163 | 0.07 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr1_+_25801648 | 0.07 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr8_-_11229523 | 0.07 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr1_-_52497834 | 0.07 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr24_-_29906904 | 0.07 |
ENSDART00000178791
|
LO018627.1
|
|
| chr24_+_12989727 | 0.07 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr13_-_37619159 | 0.07 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr21_-_293146 | 0.07 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr12_+_27537357 | 0.07 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr8_-_32805214 | 0.07 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr1_+_52560549 | 0.07 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr8_-_27687095 | 0.07 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr18_-_30504587 | 0.07 |
ENSDART00000129509
ENSDART00000028033 |
emc8
|
ER membrane protein complex subunit 8 |
| chr14_+_9456675 | 0.06 |
ENSDART00000134935
|
rell2
|
RELT-like 2 |
| chr5_+_28848870 | 0.06 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr7_+_1473929 | 0.06 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr25_+_18953575 | 0.06 |
ENSDART00000155610
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr22_+_18929412 | 0.06 |
ENSDART00000161598
ENSDART00000166650 ENSDART00000015951 ENSDART00000105392 ENSDART00000131131 |
bsg
|
basigin |
| chr15_+_37973197 | 0.06 |
ENSDART00000156661
|
si:dkey-238d18.9
|
si:dkey-238d18.9 |
| chr10_-_20065149 | 0.06 |
ENSDART00000182603
|
BX323022.1
|
|
| chr12_+_13256415 | 0.06 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr15_+_46344655 | 0.06 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr7_-_24364536 | 0.06 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr18_-_14337065 | 0.06 |
ENSDART00000135703
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr22_+_8701238 | 0.06 |
ENSDART00000106092
|
zmp:0000000652
|
zmp:0000000652 |
| chr16_-_22004402 | 0.06 |
ENSDART00000145100
ENSDART00000188217 ENSDART00000180083 |
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr21_-_43952958 | 0.06 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr10_-_44560165 | 0.06 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr5_-_29152457 | 0.06 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr12_+_30360579 | 0.06 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr12_+_30360184 | 0.06 |
ENSDART00000190718
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr11_-_18604542 | 0.06 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr13_+_25199849 | 0.06 |
ENSDART00000139209
ENSDART00000130876 |
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr20_-_33497128 | 0.06 |
ENSDART00000101974
|
erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr5_+_28857969 | 0.05 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr8_-_50981175 | 0.05 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr2_-_17115256 | 0.05 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_-_7131657 | 0.05 |
ENSDART00000175565
ENSDART00000092116 |
extl2
|
exostosin-like glycosyltransferase 2 |
| chr25_-_36044583 | 0.05 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr15_-_23647078 | 0.05 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr21_-_12749501 | 0.05 |
ENSDART00000179724
|
LO018011.1
|
|
| chr6_+_112579 | 0.05 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr16_-_31451720 | 0.05 |
ENSDART00000146886
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr11_+_44503774 | 0.05 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr12_-_33859950 | 0.05 |
ENSDART00000131181
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr2_-_27385934 | 0.05 |
ENSDART00000139886
|
toe1
|
target of EGR1, exonuclease |
| chr6_-_31224563 | 0.05 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr19_+_9174166 | 0.05 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr9_+_21146862 | 0.05 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr24_+_37361111 | 0.05 |
ENSDART00000078771
|
si:ch211-183d21.3
|
si:ch211-183d21.3 |
| chr1_+_22691256 | 0.05 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr25_+_18953756 | 0.05 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr14_+_7939398 | 0.05 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr2_-_44344321 | 0.05 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr15_-_10455438 | 0.05 |
ENSDART00000158958
ENSDART00000192971 ENSDART00000162133 ENSDART00000185071 ENSDART00000192444 ENSDART00000165668 ENSDART00000175825 |
tenm4
|
teneurin transmembrane protein 4 |
| chr7_+_30051880 | 0.04 |
ENSDART00000075609
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr7_+_34794829 | 0.04 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr19_+_7636941 | 0.04 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr14_+_7939216 | 0.04 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr10_-_7892192 | 0.04 |
ENSDART00000145480
|
smtna
|
smoothelin a |
| chr3_-_29962345 | 0.04 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr20_+_9763364 | 0.04 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| chr2_+_47471647 | 0.04 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr9_+_2020667 | 0.04 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr21_+_19418563 | 0.04 |
ENSDART00000181113
ENSDART00000080110 |
amacr
|
alpha-methylacyl-CoA racemase |
| chr8_+_7033049 | 0.04 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr3_+_24577021 | 0.04 |
ENSDART00000158449
|
sp100.3
|
SP110 nuclear body protein, tandem duplicate 3 |
| chr18_+_14277003 | 0.04 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr18_-_26797723 | 0.04 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr22_+_10158502 | 0.04 |
ENSDART00000005869
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr25_-_9013963 | 0.04 |
ENSDART00000073912
|
rag2
|
recombination activating gene 2 |
| chr18_-_444168 | 0.04 |
ENSDART00000170389
|
wtip
|
WT1 interacting protein |
| chr11_+_21910343 | 0.04 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr8_+_25247245 | 0.04 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr18_+_48802154 | 0.04 |
ENSDART00000191403
|
bmp16
|
bone morphogenetic protein 16 |
| chr21_+_21357309 | 0.04 |
ENSDART00000079634
ENSDART00000146102 |
kcnk12l
|
potassium channel, subfamily K, member 12 like |
| chr20_+_42668875 | 0.04 |
ENSDART00000048890
|
slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr16_-_28727763 | 0.04 |
ENSDART00000149575
|
dcst1
|
DC-STAMP domain containing 1 |
| chr15_+_22435460 | 0.04 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr10_-_31805923 | 0.04 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr1_-_31140096 | 0.04 |
ENSDART00000172243
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr7_-_66864756 | 0.04 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr1_-_55118745 | 0.04 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr5_+_28830388 | 0.04 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr5_+_13549603 | 0.04 |
ENSDART00000171270
|
ckap2l
|
cytoskeleton associated protein 2-like |
| chr23_-_17450746 | 0.04 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr21_-_20329541 | 0.04 |
ENSDART00000166049
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr5_+_58455488 | 0.04 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr18_+_8901846 | 0.04 |
ENSDART00000132109
ENSDART00000144247 |
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr18_+_34861568 | 0.03 |
ENSDART00000192825
|
LO018333.1
|
|
| chr10_+_22891126 | 0.03 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr24_+_2495197 | 0.03 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr8_-_19280856 | 0.03 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr12_-_26851726 | 0.03 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr14_+_21783229 | 0.03 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr8_-_18367203 | 0.03 |
ENSDART00000190744
|
dmap1
|
DNA methyltransferase 1 associated protein 1 |
| chr2_+_30480907 | 0.03 |
ENSDART00000041378
ENSDART00000138863 |
fam173b
|
family with sequence similarity 173, member B |
| chr15_-_18197008 | 0.03 |
ENSDART00000147632
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr24_+_3050020 | 0.03 |
ENSDART00000004241
|
inhbaa
|
inhibin, beta Aa |
| chr6_+_41452979 | 0.03 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr8_-_44868020 | 0.03 |
ENSDART00000142712
|
cacna1fa
|
calcium channel, voltage-dependent, L type, alpha 1F subunit a |
| chr2_+_47471943 | 0.03 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr2_-_31833347 | 0.03 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr6_+_23809501 | 0.03 |
ENSDART00000168701
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr17_-_14836320 | 0.03 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr3_-_40836081 | 0.03 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr15_-_727479 | 0.03 |
ENSDART00000157279
|
si:dkey-7i4.21
|
si:dkey-7i4.21 |
| chr2_+_32016256 | 0.03 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr7_-_19369002 | 0.03 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr11_-_44999858 | 0.03 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr2_+_16160906 | 0.03 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr7_-_17690756 | 0.03 |
ENSDART00000173759
|
map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr21_-_39253647 | 0.03 |
ENSDART00000133034
|
tusc5b
|
tumor suppressor candidate 5b |
| chr23_-_31346319 | 0.03 |
ENSDART00000008401
ENSDART00000138106 |
phip
|
pleckstrin homology domain interacting protein |
| chr6_-_43007264 | 0.03 |
ENSDART00000124623
|
si:ch73-361p23.3
|
si:ch73-361p23.3 |
| chr18_-_14337450 | 0.03 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr13_+_24584401 | 0.03 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr25_+_3326885 | 0.03 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr13_+_28612313 | 0.03 |
ENSDART00000077383
|
borcs7
|
BLOC-1 related complex subunit 7 |
| chr11_+_6136220 | 0.03 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr5_+_1911814 | 0.03 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr25_-_37160606 | 0.03 |
ENSDART00000152240
|
si:ch1073-174d20.2
|
si:ch1073-174d20.2 |
| chr21_-_22357985 | 0.02 |
ENSDART00000101751
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr23_-_44470253 | 0.02 |
ENSDART00000176333
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr15_-_16098531 | 0.02 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr14_+_21783400 | 0.02 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr4_-_815871 | 0.02 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr3_+_58379450 | 0.02 |
ENSDART00000155759
|
sdr42e2
|
short chain dehydrogenase/reductase family 42E, member 2 |
| chr16_+_47428721 | 0.02 |
ENSDART00000180597
ENSDART00000032309 |
slc25a32b
|
solute carrier family 25 (mitochondrial folate carrier), member 32b |
| chr21_-_32467099 | 0.02 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr12_+_27243059 | 0.02 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr6_-_54348568 | 0.02 |
ENSDART00000156501
|
zgc:101577
|
zgc:101577 |
| chr15_+_42599501 | 0.02 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr24_-_37338739 | 0.02 |
ENSDART00000146844
|
tsr3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf21
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.2 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.0 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.0 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.0 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0097519 | DNA recombinase complex(GO:0097519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.0 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.0 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |