Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
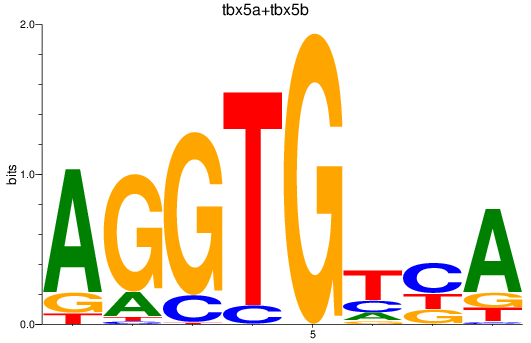
Results for tbx5a+tbx5b
Z-value: 0.43
Transcription factors associated with tbx5a+tbx5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx5a
|
ENSDARG00000024894 | T-box transcription factor 5a |
|
tbx5b
|
ENSDARG00000092060 | T-box transcription factor 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx5a | dr11_v1_chr5_-_72289648_72289648 | 0.57 | 1.1e-01 | Click! |
| tbx5b | dr11_v1_chr5_+_23152282_23152282 | -0.52 | 1.5e-01 | Click! |
Activity profile of tbx5a+tbx5b motif
Sorted Z-values of tbx5a+tbx5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_34999379 | 1.20 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr19_-_34995629 | 0.92 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr24_-_10021341 | 0.86 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr24_-_10006158 | 0.83 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr24_-_9997948 | 0.77 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr10_+_19596214 | 0.72 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr24_+_10027902 | 0.68 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr24_+_15655233 | 0.66 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr11_+_29537756 | 0.66 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr10_+_19569052 | 0.62 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr20_-_43743700 | 0.61 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr11_-_6452444 | 0.59 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr10_+_19595009 | 0.57 |
ENSDART00000112276
|
zgc:173837
|
zgc:173837 |
| chr22_+_26600834 | 0.57 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr20_-_38778479 | 0.56 |
ENSDART00000185599
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr18_+_16053455 | 0.55 |
ENSDART00000189163
ENSDART00000188269 |
FO834850.1
|
|
| chr7_-_55648336 | 0.53 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr2_-_38114370 | 0.53 |
ENSDART00000131837
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr4_-_77114795 | 0.53 |
ENSDART00000144849
|
CU467646.2
|
|
| chr24_+_15655069 | 0.52 |
ENSDART00000042943
ENSDART00000181296 |
fbxo15
|
F-box protein 15 |
| chr6_-_25165693 | 0.51 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr15_+_20530649 | 0.47 |
ENSDART00000186312
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr3_+_43086548 | 0.45 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr10_-_21542702 | 0.45 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr10_+_40324395 | 0.45 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr23_-_29812667 | 0.44 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr5_-_16475374 | 0.44 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr16_+_3982590 | 0.43 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr7_-_69795488 | 0.43 |
ENSDART00000162414
|
USP53 (1 of many)
|
ubiquitin specific peptidase 53 |
| chr20_-_14114078 | 0.43 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr8_-_1838315 | 0.42 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr17_-_16324565 | 0.42 |
ENSDART00000030835
|
hmbox1a
|
homeobox containing 1a |
| chr6_+_54498220 | 0.41 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr1_-_6085750 | 0.41 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr5_-_68333081 | 0.40 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr7_-_20021450 | 0.40 |
ENSDART00000182363
|
trip6
|
thyroid hormone receptor interactor 6 |
| chr21_-_30254185 | 0.40 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr11_-_25853212 | 0.39 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr8_-_25569920 | 0.39 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr10_-_45029041 | 0.39 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr22_-_38258053 | 0.38 |
ENSDART00000132516
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr18_-_14274803 | 0.38 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr2_+_30531726 | 0.38 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr3_+_58472305 | 0.37 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr13_+_47821524 | 0.37 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr5_+_57658898 | 0.36 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr25_+_11281970 | 0.36 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr9_-_32912638 | 0.36 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr3_+_35542067 | 0.36 |
ENSDART00000146529
ENSDART00000084549 |
rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr9_-_28939181 | 0.36 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr22_-_17653143 | 0.35 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr3_-_60027255 | 0.35 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr7_+_38962207 | 0.35 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr19_-_6083761 | 0.34 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr5_+_13472234 | 0.34 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr3_+_17456428 | 0.34 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr2_+_5843694 | 0.34 |
ENSDART00000182364
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr8_+_3431671 | 0.34 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr22_-_17652914 | 0.34 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr11_-_43473824 | 0.34 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr5_+_29652513 | 0.34 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr1_-_40341306 | 0.33 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr3_-_42016693 | 0.33 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr13_-_28610965 | 0.33 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr5_+_6670945 | 0.33 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr7_+_15329819 | 0.33 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr7_-_54217547 | 0.33 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr5_-_51191351 | 0.32 |
ENSDART00000141424
|
snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr15_+_40079468 | 0.32 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr24_+_8904135 | 0.32 |
ENSDART00000066782
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr22_+_24603930 | 0.32 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr25_-_4530085 | 0.32 |
ENSDART00000144907
|
pidd1
|
p53-induced death domain protein 1 |
| chr16_+_50434668 | 0.31 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr21_-_3700334 | 0.31 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr23_-_10745288 | 0.31 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr19_-_8812891 | 0.31 |
ENSDART00000151134
ENSDART00000025385 ENSDART00000180291 |
cers2a
|
ceramide synthase 2a |
| chr15_-_30714130 | 0.31 |
ENSDART00000156914
ENSDART00000154714 |
msi2b
|
musashi RNA-binding protein 2b |
| chr7_+_48297842 | 0.30 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr3_+_49074008 | 0.30 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr16_-_42750295 | 0.30 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr1_+_19602389 | 0.30 |
ENSDART00000088933
ENSDART00000141579 ENSDART00000111555 |
fbxo10
|
F-box protein 10 |
| chr23_+_19790962 | 0.30 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr15_-_25269028 | 0.30 |
ENSDART00000078230
ENSDART00000193872 |
mettl16
|
methyltransferase like 16 |
| chr7_+_15313443 | 0.30 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr3_+_40255408 | 0.30 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr5_+_26213874 | 0.29 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr16_-_41762983 | 0.29 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr3_+_41726360 | 0.29 |
ENSDART00000154401
|
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr13_+_40501455 | 0.29 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr8_+_9715010 | 0.29 |
ENSDART00000139414
|
gripap1
|
GRIP1 associated protein 1 |
| chr16_+_30316177 | 0.29 |
ENSDART00000109505
|
tmem79b
|
transmembrane protein 79b |
| chr21_-_35325466 | 0.28 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr3_+_15776446 | 0.28 |
ENSDART00000146651
|
znf652
|
zinc finger protein 652 |
| chr13_-_34858500 | 0.28 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr3_+_60607241 | 0.28 |
ENSDART00000167512
|
mfsd11
|
major facilitator superfamily domain containing 11 |
| chr5_-_33215261 | 0.28 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr3_-_40232615 | 0.28 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr2_-_23390779 | 0.28 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr10_+_34001444 | 0.27 |
ENSDART00000149934
|
kl
|
klotho |
| chr10_+_6884123 | 0.27 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr21_+_39197418 | 0.27 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr17_+_15674052 | 0.27 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr4_+_16768961 | 0.26 |
ENSDART00000143926
|
BX649500.1
|
|
| chr10_+_16501699 | 0.26 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr14_+_7699443 | 0.26 |
ENSDART00000123139
|
brd8
|
bromodomain containing 8 |
| chr20_-_54075136 | 0.26 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr10_+_6010570 | 0.26 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr5_-_31856681 | 0.26 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr21_+_39197628 | 0.26 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr18_+_36786842 | 0.26 |
ENSDART00000123264
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr5_-_16475682 | 0.26 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr15_-_25365319 | 0.26 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr2_-_13691834 | 0.26 |
ENSDART00000186570
|
CABZ01044235.1
|
|
| chr10_+_11038192 | 0.26 |
ENSDART00000047954
|
ugcg
|
UDP-glucose ceramide glucosyltransferase |
| chr3_-_54607166 | 0.25 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr10_-_1961930 | 0.25 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr9_+_17982737 | 0.25 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr2_-_23391266 | 0.25 |
ENSDART00000159048
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr13_-_50341004 | 0.25 |
ENSDART00000038120
|
cacul1
|
CDK2 associated cullin domain 1 |
| chr5_-_32396929 | 0.25 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr12_+_29236274 | 0.25 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr19_+_791538 | 0.25 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr15_-_1590858 | 0.25 |
ENSDART00000081875
|
nnr
|
nanor |
| chr11_-_33868881 | 0.25 |
ENSDART00000163295
ENSDART00000172633 ENSDART00000171439 |
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr3_+_15773991 | 0.24 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr2_-_32237916 | 0.24 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr10_+_39304422 | 0.24 |
ENSDART00000019267
|
kcnj1b
|
potassium inwardly-rectifying channel, subfamily J, member 1b |
| chr5_-_32338866 | 0.24 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_-_36727462 | 0.24 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr7_+_40081630 | 0.24 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr22_+_15959844 | 0.24 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr13_-_51846224 | 0.24 |
ENSDART00000184663
|
LT631684.2
|
|
| chr23_+_42254960 | 0.24 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr15_-_45538773 | 0.24 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr1_+_49814461 | 0.24 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr23_-_20175674 | 0.24 |
ENSDART00000112840
ENSDART00000189562 |
usp19
|
ubiquitin specific peptidase 19 |
| chr10_-_35149513 | 0.23 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr7_+_38962459 | 0.23 |
ENSDART00000173851
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr23_-_40194732 | 0.23 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr21_+_37357578 | 0.23 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr16_+_10264286 | 0.23 |
ENSDART00000091377
|
mrs2
|
MRS2 magnesium transporter |
| chr17_+_10570906 | 0.23 |
ENSDART00000182371
ENSDART00000176097 ENSDART00000110593 ENSDART00000169356 |
mgaa
|
MGA, MAX dimerization protein a |
| chr6_-_39199070 | 0.23 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr12_+_19976400 | 0.23 |
ENSDART00000153177
|
mkl2a
|
MKL/myocardin-like 2a |
| chr24_-_2423791 | 0.23 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr23_+_39963599 | 0.23 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr8_-_19051906 | 0.23 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr21_-_5831413 | 0.22 |
ENSDART00000150914
|
wu:fj64h06
|
wu:fj64h06 |
| chr11_-_40504170 | 0.22 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr9_+_46745348 | 0.22 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr16_+_10264601 | 0.22 |
ENSDART00000186167
|
mrs2
|
MRS2 magnesium transporter |
| chr23_+_2825940 | 0.22 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr23_+_34321237 | 0.22 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr19_-_10295173 | 0.22 |
ENSDART00000136697
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr13_-_8304605 | 0.22 |
ENSDART00000080460
|
atl2
|
atlastin GTPase 2 |
| chr4_+_14981854 | 0.22 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr8_-_13269300 | 0.22 |
ENSDART00000014528
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr4_+_20812900 | 0.22 |
ENSDART00000005847
|
nav3
|
neuron navigator 3 |
| chr6_-_51665454 | 0.22 |
ENSDART00000028607
|
chd6
|
chromodomain helicase DNA binding protein 6 |
| chr18_+_45666489 | 0.22 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr21_-_2287589 | 0.22 |
ENSDART00000161554
|
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr18_+_15841449 | 0.22 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr15_-_16177603 | 0.22 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr4_-_65323874 | 0.22 |
ENSDART00000175271
ENSDART00000187913 |
BX571813.1
|
|
| chr5_+_31350927 | 0.21 |
ENSDART00000028237
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr19_+_33850705 | 0.21 |
ENSDART00000160356
|
pex1
|
peroxisomal biogenesis factor 1 |
| chr21_+_21010786 | 0.21 |
ENSDART00000079692
|
ndr1
|
nodal-related 1 |
| chr24_+_26337623 | 0.21 |
ENSDART00000145637
|
mynn
|
myoneurin |
| chr2_-_32592084 | 0.21 |
ENSDART00000184752
|
fastk
|
Fas-activated serine/threonine kinase |
| chr10_-_28519505 | 0.21 |
ENSDART00000137964
|
bbx
|
bobby sox homolog (Drosophila) |
| chr10_+_36441124 | 0.21 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr11_+_3501669 | 0.21 |
ENSDART00000160808
|
pusl1
|
pseudouridylate synthase-like 1 |
| chr16_-_12097394 | 0.21 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr17_-_10059557 | 0.20 |
ENSDART00000092209
ENSDART00000161243 |
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr10_-_15849027 | 0.20 |
ENSDART00000184682
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr19_+_37135700 | 0.20 |
ENSDART00000103159
|
smim12
|
small integral membrane protein 12 |
| chr7_+_41887429 | 0.20 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr23_-_32156278 | 0.20 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr19_-_5812319 | 0.20 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr11_-_18015534 | 0.20 |
ENSDART00000181953
|
qrich1
|
glutamine-rich 1 |
| chr5_+_68807170 | 0.20 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr24_+_17349177 | 0.20 |
ENSDART00000176338
|
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr2_-_58075414 | 0.20 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr11_+_5588122 | 0.20 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr16_-_30878521 | 0.20 |
ENSDART00000141403
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr9_+_27720428 | 0.19 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr23_-_27505825 | 0.19 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr4_-_8093753 | 0.19 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr15_-_43164591 | 0.19 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr13_+_22717939 | 0.19 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr15_-_1622468 | 0.19 |
ENSDART00000149008
ENSDART00000034456 |
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr12_-_5448993 | 0.19 |
ENSDART00000181802
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr12_-_31457301 | 0.19 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr15_-_23442891 | 0.19 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr3_+_52953489 | 0.19 |
ENSDART00000125136
|
dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr20_+_1316803 | 0.19 |
ENSDART00000152586
ENSDART00000152165 |
nup43
|
nucleoporin 43 |
| chr8_-_38201415 | 0.19 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr21_-_21178410 | 0.19 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr13_+_1381953 | 0.18 |
ENSDART00000019983
|
rab23
|
RAB23, member RAS oncogene family |
| chr9_-_53537989 | 0.18 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx5a+tbx5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.0 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.3 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.1 | 0.5 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.3 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.3 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.3 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.4 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.5 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.2 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.3 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.2 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.4 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.4 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.2 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.4 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.6 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.5 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.4 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.5 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.3 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.2 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.5 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.2 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.2 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 0.4 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |