Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for tbx20
Z-value: 0.27
Transcription factors associated with tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx20
|
ENSDARG00000005150 | T-box transcription factor 20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx20 | dr11_v1_chr16_-_42523744_42523744 | 0.29 | 4.6e-01 | Click! |
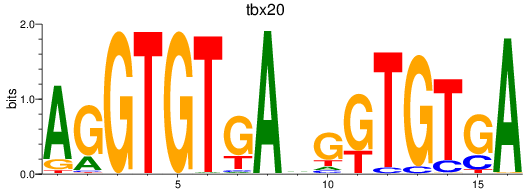
Activity profile of tbx20 motif
Sorted Z-values of tbx20 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_32228538 | 0.51 |
ENSDART00000077471
|
myhc4
|
myosin heavy chain 4 |
| chr1_-_59360114 | 0.50 |
ENSDART00000160445
|
CABZ01052573.1
|
|
| chr21_+_30351256 | 0.49 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr3_-_5964557 | 0.42 |
ENSDART00000184738
|
BX284638.1
|
|
| chr25_+_36152215 | 0.35 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr21_-_2273244 | 0.35 |
ENSDART00000171417
|
si:ch73-299h12.3
|
si:ch73-299h12.3 |
| chr9_+_23900703 | 0.32 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr4_+_47257854 | 0.32 |
ENSDART00000173868
|
crestin
|
crestin |
| chr12_-_43664682 | 0.31 |
ENSDART00000159423
|
foxi1
|
forkhead box i1 |
| chr2_+_43469241 | 0.30 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr6_+_9130989 | 0.29 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr5_+_12515402 | 0.29 |
ENSDART00000103278
|
zgc:171242
|
zgc:171242 |
| chr15_-_31516558 | 0.29 |
ENSDART00000156427
ENSDART00000156072 ENSDART00000156047 |
hmgb1b
|
high mobility group box 1b |
| chr13_-_31812394 | 0.28 |
ENSDART00000124445
|
sertad4
|
SERTA domain containing 4 |
| chr11_+_24729346 | 0.28 |
ENSDART00000087740
|
zgc:153953
|
zgc:153953 |
| chr1_-_26027327 | 0.26 |
ENSDART00000171292
ENSDART00000170878 |
si:ch211-145b13.6
|
si:ch211-145b13.6 |
| chr3_-_33417826 | 0.25 |
ENSDART00000084284
|
abi3a
|
ABI family, member 3a |
| chr19_+_26718074 | 0.24 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr7_-_69853453 | 0.22 |
ENSDART00000049928
|
myoz2a
|
myozenin 2a |
| chr7_+_39389273 | 0.21 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr24_+_13869092 | 0.21 |
ENSDART00000176492
|
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr8_-_52859301 | 0.20 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr5_-_44843738 | 0.19 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr22_-_34872533 | 0.18 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr12_+_15290800 | 0.18 |
ENSDART00000145656
|
med1
|
mediator complex subunit 1 |
| chr1_-_462165 | 0.18 |
ENSDART00000152799
|
si:ch73-244f7.3
|
si:ch73-244f7.3 |
| chr25_+_37130450 | 0.17 |
ENSDART00000183358
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr17_+_26965351 | 0.17 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr20_+_826459 | 0.16 |
ENSDART00000104740
|
nt5e
|
5'-nucleotidase, ecto (CD73) |
| chr2_-_31301929 | 0.16 |
ENSDART00000191992
ENSDART00000190723 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr14_+_20911310 | 0.15 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr7_+_38249858 | 0.15 |
ENSDART00000150158
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr8_+_68864 | 0.14 |
ENSDART00000164574
|
prr16
|
proline rich 16 |
| chr12_+_763051 | 0.14 |
ENSDART00000152579
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr7_+_756942 | 0.12 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr3_-_8285123 | 0.11 |
ENSDART00000158699
ENSDART00000138588 |
trim35-9
|
tripartite motif containing 35-9 |
| chr11_+_44622472 | 0.11 |
ENSDART00000159068
ENSDART00000166323 ENSDART00000187753 |
rbm34
|
RNA binding motif protein 34 |
| chr25_-_7686201 | 0.10 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr13_+_18276187 | 0.10 |
ENSDART00000141009
|
tet1
|
tet methylcytosine dioxygenase 1 |
| chr4_+_32156734 | 0.10 |
ENSDART00000163904
|
CR450785.1
|
|
| chr24_+_41940299 | 0.09 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr3_+_25857008 | 0.09 |
ENSDART00000154899
ENSDART00000193762 |
zgc:171844
|
zgc:171844 |
| chr24_+_42948 | 0.09 |
ENSDART00000122785
|
tmx3b
|
thioredoxin related transmembrane protein 3b |
| chr21_+_40232910 | 0.09 |
ENSDART00000075915
|
or115-11
|
odorant receptor, family F, subfamily 115, member 11 |
| chr23_+_21380079 | 0.09 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr11_-_44622478 | 0.07 |
ENSDART00000171837
|
tbce
|
tubulin folding cofactor E |
| chr3_+_52475058 | 0.07 |
ENSDART00000035867
|
si:ch211-241f5.3
|
si:ch211-241f5.3 |
| chr23_-_45049044 | 0.07 |
ENSDART00000067630
|
SH3GL2 (1 of many)
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr2_+_15776156 | 0.07 |
ENSDART00000190795
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr11_-_12104917 | 0.06 |
ENSDART00000136108
|
mrpl45
|
mitochondrial ribosomal protein L45 |
| chr5_+_9382301 | 0.06 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr10_+_11355841 | 0.05 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr16_-_563732 | 0.05 |
ENSDART00000183394
|
irx2a
|
iroquois homeobox 2a |
| chr11_-_44622240 | 0.05 |
ENSDART00000172258
ENSDART00000173340 |
tbce
|
tubulin folding cofactor E |
| chr8_-_11112058 | 0.05 |
ENSDART00000042755
|
ampd1
|
adenosine monophosphate deaminase 1 (isoform M) |
| chr5_-_49012569 | 0.05 |
ENSDART00000184690
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr4_+_35138928 | 0.05 |
ENSDART00000167934
ENSDART00000192116 ENSDART00000183689 |
si:dkey-279j5.1
|
si:dkey-279j5.1 |
| chr1_-_46326156 | 0.05 |
ENSDART00000040284
|
atp11a
|
ATPase phospholipid transporting 11A |
| chr3_-_5555500 | 0.05 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr3_+_17314997 | 0.05 |
ENSDART00000139763
|
stat5a
|
signal transducer and activator of transcription 5a |
| chr17_-_15600455 | 0.05 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr12_+_3848109 | 0.04 |
ENSDART00000144067
ENSDART00000192042 |
cabp5a
|
calcium binding protein 5a |
| chr8_+_36416285 | 0.04 |
ENSDART00000158400
|
si:busm1-194e12.8
|
si:busm1-194e12.8 |
| chr15_+_43799258 | 0.04 |
ENSDART00000122238
|
tyr
|
tyrosinase |
| chr4_+_59320243 | 0.04 |
ENSDART00000150611
|
znf1084
|
zinc finger protein 1084 |
| chr4_+_38103421 | 0.04 |
ENSDART00000164164
|
znf1016
|
zinc finger protein 1016 |
| chr20_-_31808779 | 0.04 |
ENSDART00000133788
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr21_+_5494561 | 0.03 |
ENSDART00000169728
|
ly6m3
|
lymphocyte antigen 6 family member M3 |
| chr24_+_26742226 | 0.03 |
ENSDART00000079721
|
ghsrb
|
growth hormone secretagogue receptor b |
| chr16_-_7379328 | 0.03 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr20_-_116999 | 0.03 |
ENSDART00000149581
|
ypel5
|
yippee-like 5 |
| chr4_-_75812937 | 0.03 |
ENSDART00000125096
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr12_+_49100365 | 0.03 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr25_-_35107791 | 0.02 |
ENSDART00000192819
|
zgc:165555
|
zgc:165555 |
| chr4_+_59234719 | 0.02 |
ENSDART00000170724
ENSDART00000192143 ENSDART00000109914 |
znf1086
|
zinc finger protein 1086 |
| chr18_-_51015718 | 0.02 |
ENSDART00000190698
|
LO018598.1
|
|
| chr25_+_37285737 | 0.02 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr5_-_36549024 | 0.02 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr17_+_33375469 | 0.02 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr19_+_177455 | 0.01 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr25_+_19170500 | 0.01 |
ENSDART00000182572
|
BX294110.2
|
|
| chr12_+_19362547 | 0.01 |
ENSDART00000180148
|
gspt1
|
G1 to S phase transition 1 |
| chr12_-_48127985 | 0.01 |
ENSDART00000193805
|
npffr1
|
neuropeptide FF receptor 1 |
| chr7_+_6879534 | 0.01 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr16_+_12660477 | 0.01 |
ENSDART00000016834
|
aicda
|
activation-induced cytidine deaminase |
| chr8_+_30747940 | 0.00 |
ENSDART00000098975
|
p2rx4b
|
purinergic receptor P2X, ligand-gated ion channel, 4b |
| chr17_+_33496067 | 0.00 |
ENSDART00000184724
|
pth2
|
parathyroid hormone 2 |
| chr2_+_15776408 | 0.00 |
ENSDART00000176628
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx20
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.3 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.2 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.3 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.2 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.0 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |