Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
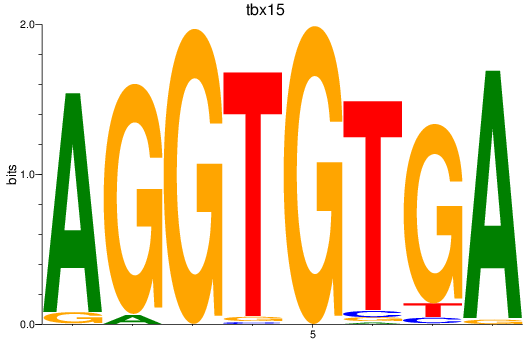
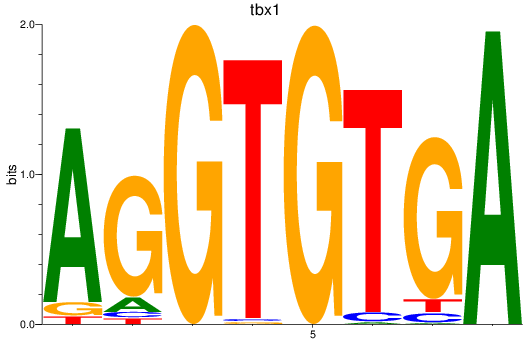
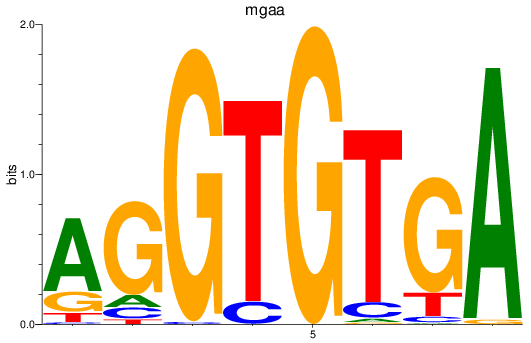
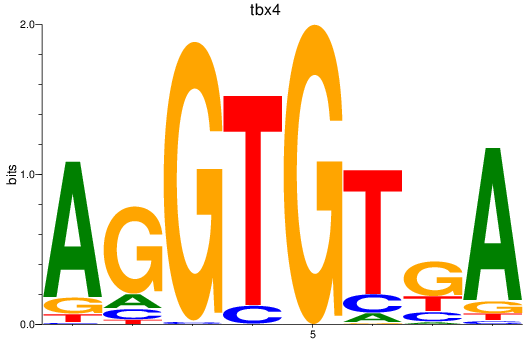
Results for tbx15_tbx1_mgaa_tbx4
Z-value: 1.66
Transcription factors associated with tbx15_tbx1_mgaa_tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx15
|
ENSDARG00000002582 | T-box transcription factor 15 |
|
tbx1
|
ENSDARG00000031891 | T-box transcription factor 1 |
|
mgaa
|
ENSDARG00000078784 | MAX dimerization protein MGA a |
|
tbx4
|
ENSDARG00000030058 | T-box transcription factor 4 |
|
tbx4
|
ENSDARG00000113067 | T-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx1 | dr11_v1_chr5_+_15202495_15202495 | 0.57 | 1.1e-01 | Click! |
| mgaa | dr11_v1_chr17_+_10566490_10566495 | -0.48 | 1.9e-01 | Click! |
| tbx15 | dr11_v1_chr9_-_21067673_21067673 | 0.37 | 3.3e-01 | Click! |
| tbx4 | dr11_v1_chr15_+_27384798_27384798 | 0.10 | 7.9e-01 | Click! |
Activity profile of tbx15_tbx1_mgaa_tbx4 motif
Sorted Z-values of tbx15_tbx1_mgaa_tbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_26965351 | 3.52 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr17_-_42568498 | 3.35 |
ENSDART00000014296
|
foxa2
|
forkhead box A2 |
| chr23_+_23232136 | 3.34 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr11_-_11575070 | 3.31 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr7_-_58776400 | 2.87 |
ENSDART00000167433
|
sox17
|
SRY (sex determining region Y)-box 17 |
| chr25_-_11016675 | 2.79 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr15_-_1590858 | 2.59 |
ENSDART00000081875
|
nnr
|
nanor |
| chr1_-_12126535 | 2.57 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr6_+_56141852 | 2.55 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr13_+_14976108 | 2.35 |
ENSDART00000011520
|
noto
|
notochord homeobox |
| chr14_-_5817039 | 2.09 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr7_-_58826164 | 2.06 |
ENSDART00000171095
|
sox32
|
SRY (sex determining region Y)-box 32 |
| chr21_+_13301798 | 2.06 |
ENSDART00000140206
|
adora2ab
|
adenosine A2a receptor b |
| chr19_-_7420867 | 2.01 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr8_-_4508248 | 1.93 |
ENSDART00000141915
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr19_+_14059349 | 1.86 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr11_+_25257022 | 1.79 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr13_+_18533005 | 1.71 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr5_+_27525477 | 1.69 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr17_-_8312923 | 1.68 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr9_+_15837398 | 1.66 |
ENSDART00000141063
|
si:dkey-103d23.5
|
si:dkey-103d23.5 |
| chr8_-_24252933 | 1.56 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr14_+_45406299 | 1.55 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr21_+_30351256 | 1.54 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr3_-_53533128 | 1.53 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr7_+_15329819 | 1.50 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr23_-_39849155 | 1.48 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr2_-_53500424 | 1.44 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr9_-_21970067 | 1.43 |
ENSDART00000009920
|
lmo7a
|
LIM domain 7a |
| chr3_-_23643751 | 1.36 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr11_+_24729346 | 1.34 |
ENSDART00000087740
|
zgc:153953
|
zgc:153953 |
| chr7_+_6969909 | 1.32 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr4_+_912563 | 1.32 |
ENSDART00000103631
|
ripply2
|
ripply transcriptional repressor 2 |
| chr7_+_7048245 | 1.29 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr19_-_5372572 | 1.26 |
ENSDART00000151326
|
krt17
|
keratin 17 |
| chr13_+_22295905 | 1.26 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr1_-_52498146 | 1.24 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr16_-_26676685 | 1.21 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr13_-_33822550 | 1.18 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr19_-_9776075 | 1.17 |
ENSDART00000140332
|
si:dkey-14o18.1
|
si:dkey-14o18.1 |
| chr22_-_12160283 | 1.16 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr7_-_34262080 | 1.15 |
ENSDART00000183246
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr7_-_52417777 | 1.14 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr16_+_53203370 | 1.12 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr8_-_11229523 | 1.11 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr8_-_14179798 | 1.11 |
ENSDART00000040645
ENSDART00000146749 |
rhoaa
|
ras homolog gene family, member Aa |
| chr12_-_43664682 | 1.09 |
ENSDART00000159423
|
foxi1
|
forkhead box i1 |
| chr4_+_65649267 | 1.08 |
ENSDART00000184068
|
si:dkey-205i10.1
|
si:dkey-205i10.1 |
| chr17_+_38255105 | 1.05 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr11_-_11910225 | 1.05 |
ENSDART00000159922
|
si:ch211-69b7.6
|
si:ch211-69b7.6 |
| chr14_-_32744464 | 1.04 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr4_-_63848305 | 1.02 |
ENSDART00000097308
|
BX901974.1
|
|
| chr10_-_31562695 | 1.01 |
ENSDART00000186456
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr12_+_15290800 | 1.01 |
ENSDART00000145656
|
med1
|
mediator complex subunit 1 |
| chr22_-_3152357 | 1.00 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr16_+_8716800 | 0.99 |
ENSDART00000124693
ENSDART00000181961 |
cabz01093075.1
|
cabz01093075.1 |
| chr5_+_34623107 | 0.98 |
ENSDART00000184126
|
enc1
|
ectodermal-neural cortex 1 |
| chr17_-_31164219 | 0.96 |
ENSDART00000055754
|
pkdccb
|
protein kinase domain containing, cytoplasmic b |
| chr14_+_6605675 | 0.95 |
ENSDART00000143179
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr4_-_20118468 | 0.95 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr14_-_26177156 | 0.95 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr3_-_40528333 | 0.93 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr3_-_32925476 | 0.93 |
ENSDART00000189673
|
aoc2
|
amine oxidase, copper containing 2 |
| chr13_+_44495063 | 0.92 |
ENSDART00000169591
|
CU694264.1
|
|
| chr20_+_34374735 | 0.91 |
ENSDART00000144090
|
si:dkeyp-11g8.3
|
si:dkeyp-11g8.3 |
| chr9_+_17438765 | 0.90 |
ENSDART00000138953
|
rgcc
|
regulator of cell cycle |
| chr20_-_43723860 | 0.89 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr5_+_37837245 | 0.89 |
ENSDART00000171617
|
epd
|
ependymin |
| chr9_+_21243985 | 0.88 |
ENSDART00000131586
|
si:rp71-68n21.12
|
si:rp71-68n21.12 |
| chr4_-_60312069 | 0.88 |
ENSDART00000167207
|
si:dkey-248e17.4
|
si:dkey-248e17.4 |
| chr13_+_33024547 | 0.88 |
ENSDART00000057377
|
arg2
|
arginase 2 |
| chr4_+_65537216 | 0.87 |
ENSDART00000181458
|
si:dkey-205i10.1
|
si:dkey-205i10.1 |
| chr3_-_34656745 | 0.87 |
ENSDART00000144824
|
znfl1
|
zinc finger-like gene 1 |
| chr21_-_2217685 | 0.87 |
ENSDART00000159315
|
si:dkey-50i6.5
|
si:dkey-50i6.5 |
| chr19_-_20148469 | 0.87 |
ENSDART00000134476
|
si:ch211-155k24.1
|
si:ch211-155k24.1 |
| chr24_-_18919562 | 0.85 |
ENSDART00000144244
ENSDART00000106188 ENSDART00000182518 |
cpa6
|
carboxypeptidase A6 |
| chr7_+_32369463 | 0.84 |
ENSDART00000180544
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr19_+_47311869 | 0.83 |
ENSDART00000136647
|
ext1c
|
exostoses (multiple) 1c |
| chr1_-_52497834 | 0.82 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr4_-_11580948 | 0.81 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr4_-_52381070 | 0.80 |
ENSDART00000139248
|
CR318589.2
|
|
| chr4_+_54519511 | 0.79 |
ENSDART00000161653
|
znf974
|
zinc finger protein 974 |
| chr9_-_31763246 | 0.79 |
ENSDART00000114742
|
si:dkey-250i3.3
|
si:dkey-250i3.3 |
| chr12_-_34887943 | 0.78 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr17_+_4408809 | 0.77 |
ENSDART00000113401
|
si:zfos-364h11.1
|
si:zfos-364h11.1 |
| chr10_-_31563049 | 0.75 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr10_-_13116337 | 0.73 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr25_-_11026907 | 0.72 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr18_+_23373683 | 0.72 |
ENSDART00000001102
ENSDART00000189030 |
mctp2a
|
multiple C2 domains, transmembrane 2a |
| chr4_-_60049792 | 0.71 |
ENSDART00000158199
|
znf1033
|
zinc finger protein 1033 |
| chr14_-_9673601 | 0.71 |
ENSDART00000188337
|
CR792437.1
|
|
| chr12_+_25600685 | 0.71 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr5_+_22791686 | 0.71 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr4_+_77740228 | 0.70 |
ENSDART00000193397
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr4_-_65747476 | 0.69 |
ENSDART00000142421
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr8_-_22508055 | 0.69 |
ENSDART00000101616
|
si:ch211-261n11.5
|
si:ch211-261n11.5 |
| chr15_+_42397125 | 0.69 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr19_-_5332784 | 0.68 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr4_-_23839789 | 0.68 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr5_-_46896541 | 0.68 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr23_-_10175898 | 0.67 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr7_+_15324830 | 0.66 |
ENSDART00000189088
|
mespaa
|
mesoderm posterior aa |
| chr17_+_58211 | 0.66 |
ENSDART00000157642
|
si:ch1073-209e23.1
|
si:ch1073-209e23.1 |
| chr16_+_40301056 | 0.65 |
ENSDART00000058578
|
rspo3
|
R-spondin 3 |
| chr13_+_4205724 | 0.64 |
ENSDART00000134105
|
dlk2
|
delta-like 2 homolog (Drosophila) |
| chr19_-_5369486 | 0.63 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr2_+_29795833 | 0.62 |
ENSDART00000056750
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr4_+_9028819 | 0.60 |
ENSDART00000102893
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr5_+_34622320 | 0.59 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr3_+_34140507 | 0.59 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr22_+_11857356 | 0.58 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr13_-_36535128 | 0.58 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr15_+_32711663 | 0.58 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr14_-_2979878 | 0.57 |
ENSDART00000031211
|
bicc2
|
bicaudal C homolog 2 |
| chr13_+_27232848 | 0.57 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr9_-_42873700 | 0.55 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr7_+_57836841 | 0.55 |
ENSDART00000136175
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr13_-_37127970 | 0.55 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr12_+_29236274 | 0.55 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr20_+_17739923 | 0.54 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr21_+_26726936 | 0.54 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr3_-_25814097 | 0.52 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr25_-_17579701 | 0.52 |
ENSDART00000073684
|
mmp15a
|
matrix metallopeptidase 15a |
| chr6_+_28877306 | 0.51 |
ENSDART00000065137
ENSDART00000123189 ENSDART00000065135 ENSDART00000181512 ENSDART00000130799 |
tp63
|
tumor protein p63 |
| chr17_+_24851951 | 0.50 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr5_-_44843738 | 0.50 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr4_-_39111612 | 0.50 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr7_-_7420301 | 0.49 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr14_-_28567845 | 0.49 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr18_+_7591381 | 0.49 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr3_+_28939759 | 0.48 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr5_+_42467867 | 0.47 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr4_+_11384891 | 0.47 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr20_+_35857399 | 0.46 |
ENSDART00000102611
|
cd2ap
|
CD2-associated protein |
| chr8_+_23245596 | 0.46 |
ENSDART00000143313
|
si:ch211-196c10.11
|
si:ch211-196c10.11 |
| chr12_-_4317165 | 0.46 |
ENSDART00000098041
|
zgc:153760
|
zgc:153760 |
| chr11_+_7264457 | 0.46 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr23_-_46020226 | 0.46 |
ENSDART00000160010
|
SYDE2
|
synapse defective Rho GTPase homolog 2 |
| chr8_+_8459192 | 0.46 |
ENSDART00000140942
ENSDART00000014939 |
comta
|
catechol-O-methyltransferase a |
| chr19_+_7735157 | 0.46 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr22_+_1300587 | 0.45 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr25_-_32751982 | 0.45 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr11_+_1796426 | 0.45 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr4_-_12323228 | 0.44 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr3_+_34121156 | 0.43 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr24_+_39158283 | 0.43 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr15_-_32131 | 0.43 |
ENSDART00000099074
ENSDART00000164323 |
CYP2C9
|
si:zfos-411a11.2 |
| chr1_+_34696503 | 0.42 |
ENSDART00000186106
|
CR339054.2
|
|
| chr19_-_32600638 | 0.42 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr3_-_61205711 | 0.42 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr15_+_32711172 | 0.41 |
ENSDART00000163936
ENSDART00000168135 |
postnb
|
periostin, osteoblast specific factor b |
| chr4_+_64562090 | 0.41 |
ENSDART00000188810
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr14_-_34771864 | 0.41 |
ENSDART00000141157
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr22_-_34872533 | 0.41 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr3_-_15451097 | 0.40 |
ENSDART00000163836
|
BX784026.1
|
Danio rerio linker for activation of T cells (lat), mRNA. |
| chr20_-_8110672 | 0.40 |
ENSDART00000113993
|
si:ch211-232i5.1
|
si:ch211-232i5.1 |
| chr7_-_51032128 | 0.40 |
ENSDART00000182781
ENSDART00000121574 |
col4a6
|
collagen, type IV, alpha 6 |
| chr12_-_16720196 | 0.40 |
ENSDART00000187639
|
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr2_+_33368414 | 0.39 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_+_36656541 | 0.39 |
ENSDART00000181349
|
si:dkey-151g22.1
|
si:dkey-151g22.1 |
| chr25_+_36327034 | 0.39 |
ENSDART00000073452
|
zgc:110216
|
zgc:110216 |
| chr12_-_16595177 | 0.39 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr2_-_47431205 | 0.38 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr20_-_7128612 | 0.38 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr22_+_16497670 | 0.38 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr7_-_38638809 | 0.38 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr7_-_48667056 | 0.37 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr8_-_28449782 | 0.36 |
ENSDART00000062702
|
cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr3_-_60288533 | 0.36 |
ENSDART00000181981
ENSDART00000193313 |
si:ch211-214b16.3
|
si:ch211-214b16.3 |
| chr13_+_45332203 | 0.36 |
ENSDART00000110466
|
si:ch211-168h21.3
|
si:ch211-168h21.3 |
| chr10_-_32524035 | 0.36 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr15_+_42599501 | 0.35 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr4_-_42221141 | 0.35 |
ENSDART00000160521
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr5_-_68795063 | 0.35 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr12_+_31616412 | 0.35 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr19_+_14921000 | 0.35 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr16_-_13921589 | 0.35 |
ENSDART00000023543
|
rcvrn2
|
recoverin 2 |
| chr19_-_5769553 | 0.34 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr11_-_30352333 | 0.34 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr10_+_21899753 | 0.34 |
ENSDART00000080155
|
hrh2b
|
histamine receptor H2b |
| chr7_-_51476276 | 0.34 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr14_-_25042184 | 0.34 |
ENSDART00000131027
|
TMEM216
|
si:rp71-1d10.5 |
| chr13_+_27232694 | 0.34 |
ENSDART00000131128
|
rin2
|
Ras and Rab interactor 2 |
| chr4_+_77735212 | 0.34 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr4_+_37952218 | 0.33 |
ENSDART00000186865
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr2_+_48288461 | 0.33 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr2_+_16453769 | 0.33 |
ENSDART00000100307
|
zgc:110269
|
zgc:110269 |
| chr24_+_22022109 | 0.33 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr4_+_64981411 | 0.33 |
ENSDART00000157798
|
CT955963.1
|
|
| chr19_+_38422059 | 0.32 |
ENSDART00000035093
|
col9a2
|
procollagen, type IX, alpha 2 |
| chr13_+_24280380 | 0.32 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr20_+_305035 | 0.31 |
ENSDART00000104807
|
si:dkey-119m7.4
|
si:dkey-119m7.4 |
| chr7_+_65673885 | 0.31 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr4_-_56151174 | 0.31 |
ENSDART00000125904
|
znf986
|
zinc finger protein 986 |
| chr19_-_22621811 | 0.31 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr5_+_54501105 | 0.31 |
ENSDART00000165889
|
tprn
|
taperin |
| chr4_-_20051141 | 0.30 |
ENSDART00000066963
|
atp6v1f
|
ATPase H+ transporting V1 subunit F |
| chr14_+_7932973 | 0.30 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr12_-_32013125 | 0.30 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr21_+_25765734 | 0.29 |
ENSDART00000021664
|
cldnb
|
claudin b |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx15_tbx1_mgaa_tbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.7 | 2.9 | GO:0048338 | axial mesoderm structural organization(GO:0048331) mesoderm structural organization(GO:0048338) |
| 0.7 | 2.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.6 | 2.5 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.6 | 3.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.5 | 1.5 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.5 | 1.4 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.5 | 6.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 1.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 2.6 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 2.1 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.2 | 3.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 2.3 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 0.6 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 2.6 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 0.6 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 1.1 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 0.5 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 1.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 1.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.8 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.5 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 1.4 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 1.7 | GO:0003128 | heart field specification(GO:0003128) |
| 0.1 | 0.8 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 0.4 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.7 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.3 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 2.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 1.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.5 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.3 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.7 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.2 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 1.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.5 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.6 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 2.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.0 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.3 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.8 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.5 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 1.0 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 1.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 1.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:1903011 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.1 | GO:0034672 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 1.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.6 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.0 | 0.2 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 2.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.8 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.3 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0002370 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of interferon-gamma production(GO:0032729) positive regulation of mast cell activation(GO:0033005) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:2000826 | regulation of heart morphogenesis(GO:2000826) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.3 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 1.6 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.2 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.0 | GO:0032925 | regulation of activin receptor signaling pathway(GO:0032925) |
| 0.0 | 0.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.0 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.9 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 1.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0042311 | vasodilation(GO:0042311) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 0.9 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.7 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.4 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.6 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 2.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 2.6 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 2.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 1.7 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 0.8 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 0.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 1.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.9 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 2.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 0.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.3 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 2.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.7 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 1.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 9.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 1.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0098918 | structural constituent of synapse(GO:0098918) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 4.3 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 1.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.9 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.1 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 3.9 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 3.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |