Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
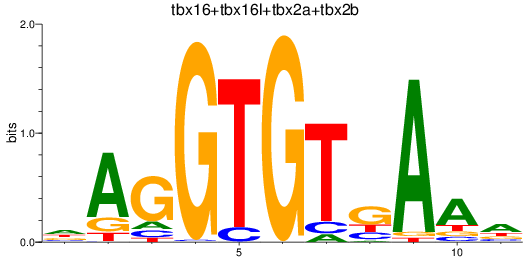
Results for tbr1b_tbx16+tbx16l+tbx2a+tbx2b
Z-value: 2.45
Transcription factors associated with tbr1b_tbx16+tbx16l+tbx2a+tbx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbr1b
|
ENSDARG00000004712 | T-box brain transcription factor 1b |
|
tbx2b
|
ENSDARG00000006120 | T-box transcription factor 2b |
|
tbx16l
|
ENSDARG00000006939 | T-box transcription factor 16, like |
|
tbx16
|
ENSDARG00000007329 | T-box transcription factor 16 |
|
tbx2a
|
ENSDARG00000018025 | T-box transcription factor 2a |
|
tbx2a
|
ENSDARG00000109541 | T-box transcription factor 2a |
|
tbx2b
|
ENSDARG00000116135 | T-box transcription factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx2b | dr11_v1_chr15_+_27364394_27364394 | -0.93 | 2.7e-04 | Click! |
| tbr1b | dr11_v1_chr9_-_51436377_51436377 | -0.93 | 3.1e-04 | Click! |
| tbx2a | dr11_v1_chr5_-_56513825_56513825 | -0.91 | 7.0e-04 | Click! |
| tbx16 | dr11_v1_chr8_-_51753604_51753604 | 0.46 | 2.1e-01 | Click! |
| tbx6l | dr11_v1_chr5_+_42280372_42280372 | -0.18 | 6.5e-01 | Click! |
Activity profile of tbr1b_tbx16+tbx16l+tbx2a+tbx2b motif
Sorted Z-values of tbr1b_tbx16+tbx16l+tbx2a+tbx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_51846224 | 7.92 |
ENSDART00000184663
|
LT631684.2
|
|
| chr1_-_49521407 | 5.27 |
ENSDART00000189845
ENSDART00000143474 |
zp3c
|
zona pellucida glycoprotein 3c |
| chr12_-_14143344 | 4.42 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr22_+_25715925 | 4.10 |
ENSDART00000150650
|
si:dkeyp-98a7.7
|
si:dkeyp-98a7.7 |
| chr22_+_25681911 | 4.06 |
ENSDART00000113381
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr22_+_25704430 | 3.98 |
ENSDART00000143776
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr19_-_34999379 | 3.88 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr22_+_25693295 | 3.86 |
ENSDART00000123888
ENSDART00000150783 |
si:dkeyp-98a7.4
si:dkeyp-98a7.3
|
si:dkeyp-98a7.4 si:dkeyp-98a7.3 |
| chr22_+_25687525 | 3.85 |
ENSDART00000135717
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr15_-_1590858 | 3.83 |
ENSDART00000081875
|
nnr
|
nanor |
| chr22_+_26600834 | 3.78 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr25_+_36292057 | 3.69 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr22_+_25720725 | 3.58 |
ENSDART00000150778
|
si:dkeyp-98a7.8
|
si:dkeyp-98a7.8 |
| chr22_+_25710148 | 3.26 |
ENSDART00000150446
|
si:dkeyp-98a7.4
|
si:dkeyp-98a7.4 |
| chr10_-_1961576 | 3.10 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr9_+_8380728 | 3.09 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr22_+_25734180 | 2.98 |
ENSDART00000143367
|
si:dkeyp-98a7.9
|
si:dkeyp-98a7.9 |
| chr10_-_1961930 | 2.94 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr8_+_3431671 | 2.84 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr20_-_14114078 | 2.69 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr21_-_3700334 | 2.67 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr16_+_15114645 | 2.64 |
ENSDART00000158483
|
mtbp
|
MDM2 binding protein |
| chr13_+_1381953 | 2.62 |
ENSDART00000019983
|
rab23
|
RAB23, member RAS oncogene family |
| chr15_-_5580093 | 2.52 |
ENSDART00000143726
|
wdr62
|
WD repeat domain 62 |
| chr15_-_17099560 | 2.36 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr6_+_54498220 | 2.34 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr5_+_25311309 | 2.30 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr2_+_30531726 | 2.29 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr13_-_9442942 | 2.29 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr23_+_42254960 | 2.29 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr11_-_25257045 | 2.25 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr2_-_31686353 | 2.25 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr2_-_6373829 | 2.15 |
ENSDART00000081633
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr11_-_11791718 | 2.14 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr2_-_38114370 | 2.13 |
ENSDART00000131837
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr19_+_791538 | 2.10 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr18_-_11595567 | 2.10 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr5_+_29652513 | 2.09 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr15_+_44184367 | 2.06 |
ENSDART00000162918
ENSDART00000110060 |
zgc:165514
|
zgc:165514 |
| chr18_-_25771553 | 2.06 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr17_-_6613458 | 2.05 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr12_-_35386910 | 2.03 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr2_-_7185460 | 2.02 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr21_+_21010786 | 1.99 |
ENSDART00000079692
|
ndr1
|
nodal-related 1 |
| chr18_+_14277003 | 1.97 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr11_-_25257595 | 1.91 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr19_+_42229018 | 1.90 |
ENSDART00000102702
|
jtb
|
jumping translocation breakpoint |
| chr1_-_21714025 | 1.89 |
ENSDART00000129066
|
zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr7_+_15329819 | 1.89 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr11_-_3535537 | 1.89 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr10_+_10972795 | 1.87 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr15_-_35112937 | 1.86 |
ENSDART00000154565
ENSDART00000099642 |
zgc:77118
|
zgc:77118 |
| chr13_-_35808904 | 1.83 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr7_+_15313443 | 1.79 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr20_-_40360571 | 1.78 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr1_-_6028876 | 1.78 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr15_-_1001177 | 1.77 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr21_+_15883546 | 1.76 |
ENSDART00000186325
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr4_+_15006217 | 1.75 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr8_-_19246342 | 1.74 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr24_-_10021341 | 1.74 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr17_+_15674052 | 1.72 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr22_-_17652112 | 1.72 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr17_-_8312923 | 1.71 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr8_+_10869183 | 1.71 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr15_-_21669618 | 1.70 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr4_-_73411863 | 1.69 |
ENSDART00000171434
|
zgc:162958
|
zgc:162958 |
| chr19_+_14109348 | 1.68 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr6_-_2133737 | 1.67 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr2_+_35733335 | 1.67 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr15_-_25269028 | 1.64 |
ENSDART00000078230
ENSDART00000193872 |
mettl16
|
methyltransferase like 16 |
| chr24_-_34680956 | 1.64 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr22_+_1300587 | 1.63 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr8_-_25605537 | 1.63 |
ENSDART00000005906
|
stk38a
|
serine/threonine kinase 38a |
| chr13_+_33368140 | 1.62 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr10_+_40324395 | 1.60 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr1_+_19602389 | 1.58 |
ENSDART00000088933
ENSDART00000141579 ENSDART00000111555 |
fbxo10
|
F-box protein 10 |
| chr19_-_34995629 | 1.58 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr15_+_44208464 | 1.54 |
ENSDART00000126868
|
CU929391.1
|
|
| chr24_+_35195963 | 1.54 |
ENSDART00000158801
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr25_+_30196039 | 1.53 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr16_+_50741154 | 1.53 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr13_-_27354003 | 1.53 |
ENSDART00000101479
ENSDART00000044652 |
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr9_-_32912638 | 1.52 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr5_+_29652198 | 1.52 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr22_-_4439311 | 1.50 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr5_-_16475682 | 1.50 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr8_+_52442622 | 1.49 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr16_-_42066523 | 1.49 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr22_-_24757785 | 1.48 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr13_+_47821524 | 1.48 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr22_-_22337382 | 1.47 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr25_+_7435291 | 1.46 |
ENSDART00000172567
ENSDART00000163017 |
prc1a
|
protein regulator of cytokinesis 1a |
| chr24_-_36593876 | 1.45 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr25_+_29662411 | 1.44 |
ENSDART00000077445
|
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr2_+_3516913 | 1.44 |
ENSDART00000109346
|
CU693445.1
|
|
| chr13_-_15793585 | 1.44 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2 associated athanogene 5 |
| chr21_-_5831413 | 1.44 |
ENSDART00000150914
|
wu:fj64h06
|
wu:fj64h06 |
| chr1_-_40341306 | 1.44 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr12_-_6880694 | 1.43 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr19_+_31532043 | 1.43 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr18_+_14329231 | 1.43 |
ENSDART00000151641
|
zgc:173742
|
zgc:173742 |
| chr6_+_59840360 | 1.42 |
ENSDART00000154647
|
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr7_-_53117131 | 1.41 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr2_+_34967210 | 1.41 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr2_+_15069011 | 1.40 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr1_+_8521323 | 1.39 |
ENSDART00000121439
ENSDART00000103626 ENSDART00000141283 |
mief2
|
mitochondrial elongation factor 2 |
| chr21_-_2185004 | 1.39 |
ENSDART00000163405
|
zgc:171220
|
zgc:171220 |
| chr1_+_30723677 | 1.39 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr1_+_30723380 | 1.38 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr17_+_23494698 | 1.37 |
ENSDART00000155150
|
kif20ba
|
kinesin family member 20Ba |
| chr19_+_33139164 | 1.37 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr18_+_36786842 | 1.36 |
ENSDART00000123264
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr11_-_40647190 | 1.36 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr12_-_10567188 | 1.35 |
ENSDART00000144283
|
myof
|
myoferlin |
| chr5_-_16475374 | 1.35 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr24_+_21174851 | 1.34 |
ENSDART00000154940
ENSDART00000155977 ENSDART00000122762 |
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr2_-_17393216 | 1.34 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr20_-_54075136 | 1.33 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr7_+_51795667 | 1.33 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr15_-_45538773 | 1.32 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr2_-_17392799 | 1.32 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr11_+_42641404 | 1.32 |
ENSDART00000172641
ENSDART00000169938 |
il17rd
|
interleukin 17 receptor D |
| chr25_-_17587785 | 1.32 |
ENSDART00000073679
ENSDART00000146851 |
zgc:66449
|
zgc:66449 |
| chr12_-_10508952 | 1.30 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr13_+_2894536 | 1.30 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr22_-_16275236 | 1.29 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr10_-_26196383 | 1.28 |
ENSDART00000192925
|
fhdc3
|
FH2 domain containing 3 |
| chr12_-_31457301 | 1.27 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr24_+_10027902 | 1.27 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr21_-_43666420 | 1.27 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr7_+_65240227 | 1.27 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr17_+_10738126 | 1.27 |
ENSDART00000051527
|
tbpl2
|
TATA box binding protein like 2 |
| chr7_-_58826164 | 1.26 |
ENSDART00000171095
|
sox32
|
SRY (sex determining region Y)-box 32 |
| chr2_+_35732652 | 1.25 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr1_-_52505279 | 1.24 |
ENSDART00000052907
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr22_+_23430688 | 1.23 |
ENSDART00000160457
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr7_+_36467796 | 1.23 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr5_+_44654535 | 1.23 |
ENSDART00000182190
ENSDART00000181872 |
dapk1
|
death-associated protein kinase 1 |
| chr8_+_52442785 | 1.23 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr23_-_27506161 | 1.22 |
ENSDART00000145007
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr7_+_15324830 | 1.22 |
ENSDART00000189088
|
mespaa
|
mesoderm posterior aa |
| chr6_-_49537646 | 1.22 |
ENSDART00000180438
|
FO704848.1
|
|
| chr8_+_9715010 | 1.22 |
ENSDART00000139414
|
gripap1
|
GRIP1 associated protein 1 |
| chr13_+_21677767 | 1.21 |
ENSDART00000165166
|
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr12_+_27231212 | 1.21 |
ENSDART00000133023
ENSDART00000123739 |
tmem106a
|
transmembrane protein 106A |
| chr25_+_25085349 | 1.21 |
ENSDART00000192166
|
si:ch73-182e20.4
|
si:ch73-182e20.4 |
| chr19_-_5812319 | 1.20 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr3_-_3439150 | 1.19 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr18_+_13077800 | 1.18 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr8_-_37101581 | 1.18 |
ENSDART00000185922
|
zgc:162200
|
zgc:162200 |
| chr20_-_37933237 | 1.18 |
ENSDART00000142567
ENSDART00000036371 ENSDART00000061445 |
angel2
|
angel homolog 2 (Drosophila) |
| chr11_+_25583950 | 1.18 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr2_-_24061575 | 1.17 |
ENSDART00000089234
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr12_-_13730501 | 1.17 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr12_+_18906407 | 1.16 |
ENSDART00000105854
|
josd1
|
Josephin domain containing 1 |
| chr22_-_14475927 | 1.16 |
ENSDART00000135768
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr2_-_39675829 | 1.16 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr25_+_32496877 | 1.15 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr14_-_35400671 | 1.15 |
ENSDART00000129676
|
lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr20_+_9124369 | 1.15 |
ENSDART00000064150
|
si:ch211-59d15.9
|
si:ch211-59d15.9 |
| chr25_+_36292465 | 1.15 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr15_+_44206698 | 1.15 |
ENSDART00000186996
|
CU929391.1
|
|
| chr5_-_29512538 | 1.15 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr6_-_19310660 | 1.15 |
ENSDART00000171110
|
sumo2a
|
small ubiquitin-like modifier 2a |
| chr17_+_34186632 | 1.14 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr23_+_42272588 | 1.14 |
ENSDART00000164907
|
CABZ01065131.1
|
|
| chr6_+_32326074 | 1.14 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr16_+_39196727 | 1.14 |
ENSDART00000017017
|
zdhhc3a
|
zinc finger, DHHC-type containing 3a |
| chr14_+_28518349 | 1.13 |
ENSDART00000159961
|
stag2b
|
stromal antigen 2b |
| chr7_+_25221757 | 1.12 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
| chr24_+_26337623 | 1.12 |
ENSDART00000145637
|
mynn
|
myoneurin |
| chr24_-_26369185 | 1.11 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr11_+_45287541 | 1.11 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr3_+_48221413 | 1.11 |
ENSDART00000170955
|
fn3krp
|
fructosamine 3 kinase related protein |
| chr23_-_893547 | 1.10 |
ENSDART00000136805
|
rbm10
|
RNA binding motif protein 10 |
| chr5_+_37406358 | 1.10 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr25_+_3507368 | 1.10 |
ENSDART00000157777
|
zgc:153293
|
zgc:153293 |
| chr5_-_28016805 | 1.10 |
ENSDART00000078642
|
vps37b
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr15_-_41689684 | 1.09 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr12_+_34854562 | 1.09 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr23_-_27505825 | 1.09 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr8_+_41048501 | 1.09 |
ENSDART00000123288
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr7_-_58776400 | 1.09 |
ENSDART00000167433
|
sox17
|
SRY (sex determining region Y)-box 17 |
| chr6_-_1432200 | 1.09 |
ENSDART00000182901
|
LO018148.1
|
|
| chr4_+_8213063 | 1.09 |
ENSDART00000027829
ENSDART00000148440 |
ERC1
|
si:dkey-222l13.1 |
| chr17_-_50059664 | 1.08 |
ENSDART00000193940
|
FO704836.1
|
|
| chr9_+_34151367 | 1.07 |
ENSDART00000143991
|
gpr161
|
G protein-coupled receptor 161 |
| chr20_+_33924235 | 1.07 |
ENSDART00000146292
ENSDART00000139609 |
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr6_-_21726758 | 1.07 |
ENSDART00000083085
|
mtmr14
|
myotubularin related protein 14 |
| chr3_+_43086548 | 1.07 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr5_-_5243079 | 1.07 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr20_+_35208020 | 1.07 |
ENSDART00000153315
ENSDART00000045135 |
fbxo16
|
F-box protein 16 |
| chr22_-_16443199 | 1.07 |
ENSDART00000006290
ENSDART00000193335 |
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr15_+_42397125 | 1.06 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr22_-_34609528 | 1.06 |
ENSDART00000190781
ENSDART00000171712 |
terf2ip
|
telomeric repeat binding factor 2, interacting protein |
| chr24_-_10393969 | 1.06 |
ENSDART00000106260
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr8_+_13364950 | 1.06 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr10_-_44411032 | 1.05 |
ENSDART00000111509
|
CABZ01072096.1
|
|
| chr5_-_69180587 | 1.05 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbr1b_tbx16+tbx16l+tbx2a+tbx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0090008 | hypoblast development(GO:0090008) |
| 1.1 | 5.6 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 1.0 | 6.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.8 | 2.4 | GO:1905132 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.7 | 2.8 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.6 | 0.6 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.6 | 1.9 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.6 | 1.8 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.6 | 1.7 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.5 | 2.8 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.5 | 1.4 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.4 | 3.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 1.3 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.4 | 6.8 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.4 | 1.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 1.3 | GO:0071962 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.3 | 2.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.3 | 1.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.3 | 2.1 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.3 | 1.5 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 1.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 1.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.3 | 1.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 1.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 1.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.3 | 1.1 | GO:1902590 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.3 | 3.6 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 1.0 | GO:0021825 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.2 | 1.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.2 | 4.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 1.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 1.2 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.2 | 1.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 3.2 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.2 | 1.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.7 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 1.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 2.0 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.2 | 0.6 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.2 | 1.9 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 0.6 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 2.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.8 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 3.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 2.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 0.8 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 2.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 1.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 1.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.2 | 1.7 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 1.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 5.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 1.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.5 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.2 | 1.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.2 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 0.3 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.2 | 0.9 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 1.8 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.2 | 0.5 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 0.5 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 1.6 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 1.3 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.2 | 0.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 1.5 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.4 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 1.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.8 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 1.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.5 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.5 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.9 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 1.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.7 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.9 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.9 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.5 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 1.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.4 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.7 | GO:0003128 | heart field specification(GO:0003128) |
| 0.1 | 1.0 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.9 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 3.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.3 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.4 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.6 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.4 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.6 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.5 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.6 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.7 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.4 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 0.3 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.5 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.2 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.1 | 2.8 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 0.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 2.7 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.1 | 0.3 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 2.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 1.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.6 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 2.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 2.0 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 0.6 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.6 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.6 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.2 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.1 | 0.7 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.4 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 1.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.3 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 3.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 2.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 2.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.1 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 0.5 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 1.9 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.8 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.5 | GO:1904375 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 0.5 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 1.3 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.2 | GO:0060251 | glial cell proliferation(GO:0014009) regulation of glial cell proliferation(GO:0060251) negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 1.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 1.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.6 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.9 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 2.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 1.2 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.0 | 1.9 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 0.4 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.2 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) skin epidermis development(GO:0098773) |
| 0.0 | 0.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 1.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0033206 | female meiotic division(GO:0007143) meiotic cytokinesis(GO:0033206) cleavage furrow formation(GO:0036089) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 1.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.3 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.4 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 1.4 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 1.8 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 0.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 1.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.7 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0042987 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 2.2 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.8 | GO:0007338 | single fertilization(GO:0007338) fertilization(GO:0009566) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 3.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0033003 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) positive regulation of B cell mediated immunity(GO:0002714) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of immunoglobulin mediated immune response(GO:0002891) positive regulation of interferon-gamma production(GO:0032729) regulation of mast cell activation(GO:0033003) positive regulation of mast cell activation(GO:0033005) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.5 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.6 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0033338 | medial fin development(GO:0033338) |
| 0.0 | 1.3 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 1.0 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 8.8 | GO:0006511 | ubiquitin-dependent protein catabolic process(GO:0006511) |
| 0.0 | 0.3 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.3 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 5.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.1 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.2 | GO:0031936 | regulation of chromatin silencing(GO:0031935) negative regulation of chromatin silencing(GO:0031936) negative regulation of gene silencing(GO:0060969) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.4 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.8 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 2.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 4.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.7 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.0 | 0.1 | GO:0098700 | equilibrioception(GO:0050957) neurotransmitter loading into synaptic vesicle(GO:0098700) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | GO:1990923 | PET complex(GO:1990923) |
| 0.7 | 3.6 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.4 | 2.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.3 | 2.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 1.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 4.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 1.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 1.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 1.6 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 0.6 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.5 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 0.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 2.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 3.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 2.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 2.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 1.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.7 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 2.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.4 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 4.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.3 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 1.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.4 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.1 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 5.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.1 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.2 | GO:0031511 | condensed chromosome inner kinetochore(GO:0000939) Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.3 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 1.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 2.0 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 3.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 4.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 2.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 5.7 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 3.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 8.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.6 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 1.1 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 5.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.7 | 8.8 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.6 | 4.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.5 | 1.4 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.4 | 1.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 6.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 2.0 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.4 | 2.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.4 | 2.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.4 | 2.5 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.3 | 1.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.3 | 1.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 1.6 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.3 | 1.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 4.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 3.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 0.7 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 0.9 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.2 | 1.6 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 4.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 1.1 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.2 | 2.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.2 | 0.6 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.2 | 1.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.2 | 0.6 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.2 | 1.9 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 2.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 0.5 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 1.0 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.2 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 1.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.4 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 3.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 2.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 31.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.5 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.3 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.6 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 1.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.3 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 1.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.0 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 1.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.3 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 1.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 2.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 2.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.7 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 4.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.4 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 2.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.9 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 3.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0051219 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.0 | 0.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 2.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 1.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 3.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.8 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 3.6 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 9.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.9 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 1.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 9.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 2.2 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 4.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 4.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.9 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.5 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 3.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.7 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 7.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |