Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
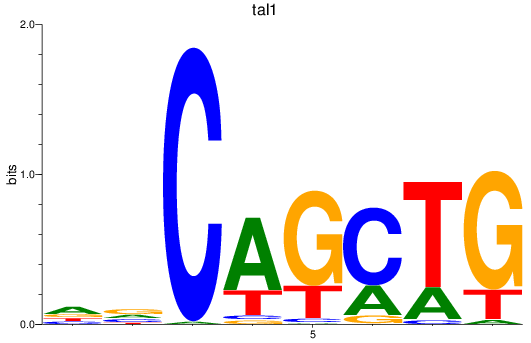
Results for tal1
Z-value: 1.45
Transcription factors associated with tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tal1
|
ENSDARG00000019930 | T-cell acute lymphocytic leukemia 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tal1 | dr11_v1_chr22_+_16535575_16535575 | 0.06 | 8.7e-01 | Click! |
Activity profile of tal1 motif
Sorted Z-values of tal1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_44715224 | 3.41 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr17_-_10073926 | 1.73 |
ENSDART00000166081
ENSDART00000161574 |
zgc:109986
|
zgc:109986 |
| chr2_-_16565690 | 1.67 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr11_+_30296332 | 1.66 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr19_-_24555935 | 1.64 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr14_-_3381303 | 1.57 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr19_-_24555623 | 1.57 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr7_+_24049776 | 1.50 |
ENSDART00000166559
|
efs
|
embryonal Fyn-associated substrate |
| chr3_-_29977495 | 1.49 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr8_-_1051438 | 1.47 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr21_+_53504 | 1.44 |
ENSDART00000170452
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr16_+_54875530 | 1.42 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr14_+_9421510 | 1.39 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr7_+_26029672 | 1.38 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr1_-_52494122 | 1.36 |
ENSDART00000131407
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr15_-_26636826 | 1.35 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr20_+_2281933 | 1.32 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr7_+_56577906 | 1.26 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr15_-_14064302 | 1.19 |
ENSDART00000156384
|
BX927244.1
|
|
| chr10_+_4987766 | 1.16 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr3_-_6767440 | 1.14 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr7_+_56577522 | 1.12 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr23_+_578218 | 1.08 |
ENSDART00000055134
|
ogfr
|
opioid growth factor receptor |
| chr4_+_8638622 | 1.01 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr2_-_36350082 | 0.99 |
ENSDART00000193104
|
CU571384.1
|
|
| chr18_-_46010 | 0.95 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr10_-_36808348 | 0.94 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr7_-_6470431 | 0.92 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr11_+_25259058 | 0.92 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr19_-_48010490 | 0.86 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr1_-_8980665 | 0.86 |
ENSDART00000148182
|
si:ch73-191k20.3
|
si:ch73-191k20.3 |
| chr4_-_48636872 | 0.82 |
ENSDART00000168605
|
znf1063
|
zinc finger protein 1063 |
| chr13_+_40770628 | 0.80 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr17_-_681142 | 0.77 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr3_+_54581987 | 0.76 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr6_-_51573975 | 0.73 |
ENSDART00000073865
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr4_+_41311345 | 0.73 |
ENSDART00000151905
|
si:dkey-16p19.8
|
si:dkey-16p19.8 |
| chr13_-_9525527 | 0.72 |
ENSDART00000190618
|
CR848040.5
|
|
| chr23_+_26079467 | 0.71 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr5_+_42467867 | 0.69 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr15_-_47857687 | 0.69 |
ENSDART00000098982
ENSDART00000151594 |
h3f3b.1
|
H3 histone, family 3B.1 |
| chr19_+_1964005 | 0.69 |
ENSDART00000172049
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr1_-_59141715 | 0.69 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr25_+_1732838 | 0.68 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr4_-_52165969 | 0.68 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr16_+_42772678 | 0.67 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr7_-_27033080 | 0.66 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr16_-_41990421 | 0.65 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr24_-_26310854 | 0.65 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr15_+_47525073 | 0.65 |
ENSDART00000067583
|
sidt2
|
SID1 transmembrane family, member 2 |
| chr14_+_17125428 | 0.63 |
ENSDART00000161489
|
slc43a3b
|
solute carrier family 43, member 3b |
| chr17_+_53156530 | 0.61 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr24_-_9300160 | 0.60 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr24_-_26399623 | 0.60 |
ENSDART00000112317
|
zgc:194621
|
zgc:194621 |
| chr1_+_54871725 | 0.59 |
ENSDART00000193654
|
si:ch211-196h16.5
|
si:ch211-196h16.5 |
| chr3_-_1232384 | 0.58 |
ENSDART00000053892
|
si:ch1073-314i13.4
|
si:ch1073-314i13.4 |
| chr2_-_55903520 | 0.58 |
ENSDART00000128828
|
cplx4c
|
complexin 4c |
| chr23_-_36934944 | 0.57 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr4_+_5249494 | 0.57 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr8_+_46327350 | 0.57 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr1_+_27868294 | 0.57 |
ENSDART00000165332
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr4_-_58964138 | 0.57 |
ENSDART00000150259
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr25_-_36369057 | 0.57 |
ENSDART00000064400
|
si:ch211-113a14.24
|
si:ch211-113a14.24 |
| chr22_-_16494406 | 0.57 |
ENSDART00000062727
|
stx6
|
syntaxin 6 |
| chr4_-_76488581 | 0.57 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr2_+_49864219 | 0.56 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr22_-_26353916 | 0.55 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr1_+_52560549 | 0.54 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr4_-_65747476 | 0.54 |
ENSDART00000142421
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr21_-_217589 | 0.53 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr4_+_38170708 | 0.52 |
ENSDART00000168900
|
znf1071
|
zinc finger protein 1071 |
| chr9_-_23747264 | 0.52 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr3_-_36115339 | 0.51 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr16_+_46807214 | 0.51 |
ENSDART00000185524
|
CABZ01027551.1
|
|
| chr22_-_28373698 | 0.51 |
ENSDART00000157592
|
si:ch211-213c4.5
|
si:ch211-213c4.5 |
| chr5_+_338154 | 0.50 |
ENSDART00000191743
|
rnf170
|
ring finger protein 170 |
| chr24_+_17269849 | 0.49 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr17_-_39786222 | 0.49 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr5_-_65000312 | 0.49 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr7_+_42935126 | 0.49 |
ENSDART00000157747
|
BX284696.1
|
|
| chr15_-_40267485 | 0.48 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr4_-_48578324 | 0.47 |
ENSDART00000122707
|
znf980
|
zinc finger protein 980 |
| chr4_-_74466703 | 0.46 |
ENSDART00000174032
ENSDART00000189417 |
LO018124.1
|
|
| chr25_+_258883 | 0.46 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr1_+_49686408 | 0.46 |
ENSDART00000140824
|
si:ch211-149l1.2
|
si:ch211-149l1.2 |
| chr25_+_35212919 | 0.45 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr14_-_36862745 | 0.45 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr13_+_22316746 | 0.45 |
ENSDART00000188968
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr16_-_41503925 | 0.44 |
ENSDART00000029492
|
cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr1_+_52563298 | 0.44 |
ENSDART00000142465
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr25_+_15933411 | 0.43 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr4_+_37952218 | 0.43 |
ENSDART00000186865
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr2_-_59157790 | 0.43 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr5_+_37785152 | 0.43 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr4_+_34126849 | 0.43 |
ENSDART00000162442
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr17_+_53311618 | 0.42 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr4_+_64562090 | 0.42 |
ENSDART00000188810
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr12_-_656540 | 0.42 |
ENSDART00000172651
|
sult2st2
|
sulfotransferase family 2, cytosolic sulfotransferase 2 |
| chr3_-_61592417 | 0.40 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr3_+_4266289 | 0.40 |
ENSDART00000101636
|
si:dkey-73p2.1
|
si:dkey-73p2.1 |
| chr10_-_8294965 | 0.40 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr10_-_43928937 | 0.40 |
ENSDART00000168014
ENSDART00000086220 |
mctp1b
|
multiple C2 domains, transmembrane 1b |
| chr22_-_10541372 | 0.39 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr3_+_3545825 | 0.39 |
ENSDART00000109060
|
CR589947.1
|
|
| chr14_+_31865099 | 0.39 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr4_-_43911462 | 0.39 |
ENSDART00000189056
ENSDART00000162441 ENSDART00000171367 |
znf1104
|
zinc finger protein 1104 |
| chr3_+_30500968 | 0.38 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr3_-_58226583 | 0.38 |
ENSDART00000187429
|
im:6904045
|
im:6904045 |
| chr2_-_30200206 | 0.38 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr17_+_10074360 | 0.38 |
ENSDART00000166649
|
srp54
|
signal recognition particle 54 |
| chr4_+_31293362 | 0.38 |
ENSDART00000169781
|
si:dkey-57l19.1
|
si:dkey-57l19.1 |
| chr2_-_59265521 | 0.38 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr3_+_58833306 | 0.37 |
ENSDART00000113223
|
igl1c3
|
immunoglobulin light 1 constant 3 |
| chr17_+_53439606 | 0.37 |
ENSDART00000154946
|
si:zfos-1714f5.3
|
si:zfos-1714f5.3 |
| chr25_-_35153985 | 0.37 |
ENSDART00000154851
|
zgc:153405
|
zgc:153405 |
| chr4_-_32456788 | 0.37 |
ENSDART00000151862
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr1_+_292545 | 0.37 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr3_-_1388936 | 0.37 |
ENSDART00000171278
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr23_-_36910656 | 0.37 |
ENSDART00000139563
|
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr4_-_67589158 | 0.37 |
ENSDART00000184361
|
BX088712.2
|
|
| chr6_+_9241121 | 0.36 |
ENSDART00000064989
|
pimr70
|
Pim proto-oncogene, serine/threonine kinase, related 70 |
| chr25_-_36044583 | 0.36 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr8_+_39570615 | 0.36 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr21_-_45073764 | 0.36 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr4_-_1497384 | 0.36 |
ENSDART00000093236
|
zmp:0000000711
|
zmp:0000000711 |
| chr16_+_19014886 | 0.35 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr5_-_67762434 | 0.35 |
ENSDART00000167301
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr3_+_5504990 | 0.35 |
ENSDART00000143626
|
si:ch73-264i18.2
|
si:ch73-264i18.2 |
| chr24_+_35827766 | 0.35 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr10_+_26896142 | 0.35 |
ENSDART00000188225
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr5_+_65946222 | 0.34 |
ENSDART00000190969
ENSDART00000161578 |
mymk
|
myomaker, myoblast fusion factor |
| chr7_+_5964296 | 0.34 |
ENSDART00000173380
|
si:dkey-23a13.17
|
si:dkey-23a13.17 |
| chr3_-_10970502 | 0.34 |
ENSDART00000127500
|
CR382337.1
|
|
| chr2_+_13694450 | 0.34 |
ENSDART00000077259
ENSDART00000189485 |
ebna1bp2
|
EBNA1 binding protein 2 |
| chr17_-_24838927 | 0.34 |
ENSDART00000123259
|
CR388164.1
|
|
| chr16_-_19568795 | 0.33 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_+_55452892 | 0.33 |
ENSDART00000122508
|
CR788255.1
|
|
| chr4_-_76129431 | 0.32 |
ENSDART00000162809
|
si:ch211-106j21.2
|
si:ch211-106j21.2 |
| chr21_-_45382112 | 0.32 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr25_-_18470695 | 0.32 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr21_-_23331619 | 0.32 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr8_+_22491947 | 0.32 |
ENSDART00000125805
|
si:ch211-261n11.8
|
si:ch211-261n11.8 |
| chr4_-_2052687 | 0.32 |
ENSDART00000138291
ENSDART00000150844 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr2_+_20793982 | 0.32 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr11_-_26040594 | 0.32 |
ENSDART00000144115
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr1_-_59232267 | 0.32 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr11_+_6136220 | 0.31 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr7_-_6346859 | 0.31 |
ENSDART00000172913
|
si:ch73-368j24.11
|
si:ch73-368j24.11 |
| chr1_+_52130213 | 0.31 |
ENSDART00000018817
|
rnf11a
|
ring finger protein 11a |
| chr11_-_8782871 | 0.31 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr20_-_31075972 | 0.31 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr25_-_3549321 | 0.31 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr14_-_25928899 | 0.30 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr10_+_40633990 | 0.30 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr10_-_5020378 | 0.30 |
ENSDART00000191857
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr2_-_59345920 | 0.30 |
ENSDART00000134662
|
ftr37
|
finTRIM family, member 37 |
| chr10_-_8295294 | 0.30 |
ENSDART00000075412
ENSDART00000163803 |
plpp1a
|
phospholipid phosphatase 1a |
| chr23_-_16682186 | 0.30 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr24_-_6029314 | 0.30 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr4_-_38033800 | 0.29 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr18_+_48802154 | 0.29 |
ENSDART00000191403
|
bmp16
|
bone morphogenetic protein 16 |
| chr7_+_33132074 | 0.29 |
ENSDART00000073554
|
zgc:153219
|
zgc:153219 |
| chr1_-_206208 | 0.29 |
ENSDART00000060968
|
adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr19_-_43639331 | 0.29 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr24_-_36337395 | 0.29 |
ENSDART00000154402
|
si:ch211-40k21.9
|
si:ch211-40k21.9 |
| chr5_-_26181863 | 0.28 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr19_+_43978814 | 0.28 |
ENSDART00000102314
|
nkd3
|
naked cuticle homolog 3 |
| chr9_-_1499721 | 0.28 |
ENSDART00000062940
|
si:ch211-222f23.6
|
si:ch211-222f23.6 |
| chr7_-_50917255 | 0.28 |
ENSDART00000022918
|
ankrd46b
|
ankyrin repeat domain 46b |
| chr4_+_50401133 | 0.28 |
ENSDART00000150644
|
si:dkey-156k2.7
|
si:dkey-156k2.7 |
| chr21_-_5007109 | 0.28 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr18_-_20560007 | 0.28 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr2_-_42492201 | 0.27 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr2_-_59247811 | 0.27 |
ENSDART00000141384
|
ftr32
|
finTRIM family, member 32 |
| chr14_-_9281232 | 0.27 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr20_-_26929685 | 0.27 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr24_-_6038025 | 0.27 |
ENSDART00000077819
ENSDART00000139216 |
ftr61
|
finTRIM family, member 61 |
| chr17_+_132555 | 0.27 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr12_+_18533198 | 0.27 |
ENSDART00000189729
|
meiob
|
meiosis specific with OB domains |
| chr25_-_173165 | 0.26 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr24_+_38155830 | 0.26 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr12_-_45238759 | 0.26 |
ENSDART00000154859
|
trim65
|
tripartite motif containing 65 |
| chr25_+_37290206 | 0.26 |
ENSDART00000086474
|
si:dkey-234i14.12
|
si:dkey-234i14.12 |
| chr3_+_3598555 | 0.26 |
ENSDART00000191152
|
CR589947.3
|
|
| chr19_+_31404686 | 0.26 |
ENSDART00000078459
|
pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr3_-_62380146 | 0.25 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr21_+_22840246 | 0.25 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr18_-_8398972 | 0.25 |
ENSDART00000136512
|
BEND7
|
si:ch211-220f12.4 |
| chr7_+_6317866 | 0.25 |
ENSDART00000173397
|
si:ch211-220f21.3
|
si:ch211-220f21.3 |
| chr25_+_3318192 | 0.25 |
ENSDART00000146154
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr18_-_5248365 | 0.25 |
ENSDART00000082506
ENSDART00000082504 ENSDART00000097960 |
myef2
|
myelin expression factor 2 |
| chr3_-_5413018 | 0.25 |
ENSDART00000063138
|
vmo1a
|
vitelline membrane outer layer 1 homolog a |
| chr4_+_61171310 | 0.25 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr14_-_22108718 | 0.25 |
ENSDART00000054410
|
med19a
|
mediator complex subunit 19a |
| chr1_+_2260407 | 0.24 |
ENSDART00000058876
|
kpnb3
|
karyopherin (importin) beta 3 |
| chr1_-_513762 | 0.24 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr7_+_25053331 | 0.24 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr3_-_3939785 | 0.24 |
ENSDART00000049593
|
unm_sa1506
|
un-named sa1506 |
| chr14_-_32258759 | 0.24 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr20_+_27020201 | 0.24 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
Network of associatons between targets according to the STRING database.
First level regulatory network of tal1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.3 | 1.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.0 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.2 | 1.0 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.7 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 1.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.5 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 2.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.3 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.3 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 3.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.8 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.3 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.2 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.5 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.7 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.3 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.2 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.7 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.8 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.3 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.7 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 1.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.4 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.2 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 0.7 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 3.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.7 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.8 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 2.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.6 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 0.7 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.2 | 1.0 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 3.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.7 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.9 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.7 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.0 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |