Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
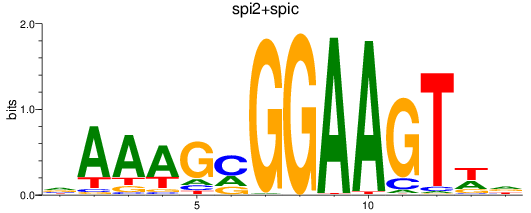
Results for spi2+spic
Z-value: 1.48
Transcription factors associated with spi2+spic
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
spic
|
ENSDARG00000012435 | Spi-C transcription factor (Spi-1/PU.1 related) |
|
spi2
|
ENSDARG00000087438 | Spi-2 proto-oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| spi2 | dr11_v1_chr4_-_17803784_17803784 | -0.70 | 3.4e-02 | Click! |
| spic | dr11_v1_chr4_-_17812131_17812131 | -0.33 | 3.8e-01 | Click! |
Activity profile of spi2+spic motif
Sorted Z-values of spi2+spic motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_38287987 | 1.90 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr4_-_17263210 | 1.88 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr9_+_8408054 | 1.80 |
ENSDART00000144373
|
zgc:153499
|
zgc:153499 |
| chr23_+_24989387 | 1.80 |
ENSDART00000172299
ENSDART00000145307 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr13_+_31716820 | 1.61 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr12_-_33354409 | 1.57 |
ENSDART00000178515
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_+_8407778 | 1.50 |
ENSDART00000102754
ENSDART00000178144 |
zgc:153499
|
zgc:153499 |
| chr12_+_13091842 | 1.41 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr15_+_34939906 | 1.32 |
ENSDART00000131182
|
si:ch73-95l15.3
|
si:ch73-95l15.3 |
| chr9_-_35633827 | 1.32 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr15_+_34940098 | 1.27 |
ENSDART00000099703
|
si:ch73-95l15.3
|
si:ch73-95l15.3 |
| chr16_-_46393154 | 1.26 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr21_+_3244146 | 1.20 |
ENSDART00000127740
|
ctif
|
CBP80/20-dependent translation initiation factor |
| chr7_+_19600262 | 1.17 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr8_-_49495584 | 1.16 |
ENSDART00000141691
|
opn7d
|
opsin 7, group member d |
| chr16_+_20934353 | 1.13 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr14_-_16810401 | 1.13 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr20_-_52928541 | 1.11 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr16_-_41488023 | 1.10 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr22_-_11829436 | 1.07 |
ENSDART00000126784
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr15_-_18209672 | 1.06 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr13_-_18695289 | 1.03 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr22_+_24623936 | 1.01 |
ENSDART00000160924
|
mcoln2
|
mucolipin 2 |
| chr21_-_9769500 | 1.01 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr5_-_16475682 | 1.00 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr4_-_14191717 | 0.98 |
ENSDART00000147928
|
pus7l
|
pseudouridylate synthase 7-like |
| chr15_-_34934784 | 0.97 |
ENSDART00000190848
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr8_+_8643901 | 0.97 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr2_-_37462462 | 0.97 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr19_-_11431278 | 0.95 |
ENSDART00000109889
|
hsbp1l1
|
heat shock factor binding protein 1 like 1 |
| chr18_+_45862414 | 0.94 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr16_-_54919260 | 0.94 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr4_-_14191434 | 0.93 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr2_-_51507540 | 0.93 |
ENSDART00000166605
ENSDART00000161093 |
pigrl2.3
|
polymeric immunoglobulin receptor-like 2.3 |
| chr1_+_44053641 | 0.92 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr4_-_17353100 | 0.91 |
ENSDART00000134467
ENSDART00000189019 |
parpbp
|
PARP1 binding protein |
| chr5_-_38248347 | 0.90 |
ENSDART00000084917
ENSDART00000139479 |
slc12a9
|
solute carrier family 12, member 9 |
| chr17_-_10059557 | 0.90 |
ENSDART00000092209
ENSDART00000161243 |
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr13_+_11829072 | 0.89 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr24_+_39034090 | 0.89 |
ENSDART00000185763
|
capn15
|
calpain 15 |
| chr14_-_24110251 | 0.89 |
ENSDART00000079226
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr15_+_34946779 | 0.88 |
ENSDART00000192661
ENSDART00000188800 ENSDART00000156515 |
si:ch73-95l15.5
zgc:55621
|
si:ch73-95l15.5 zgc:55621 |
| chr14_-_16807206 | 0.88 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr20_-_49889111 | 0.87 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr21_+_76739 | 0.86 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr20_+_2642855 | 0.86 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr17_+_39790388 | 0.84 |
ENSDART00000149488
|
ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr16_-_41487589 | 0.84 |
ENSDART00000188115
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr15_+_34934568 | 0.83 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr10_+_5954787 | 0.82 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr21_+_18907102 | 0.82 |
ENSDART00000160185
ENSDART00000190175 ENSDART00000017937 ENSDART00000191546 ENSDART00000130519 ENSDART00000137143 |
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr7_+_39006837 | 0.82 |
ENSDART00000173735
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr1_+_26105141 | 0.82 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr19_+_43669122 | 0.79 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr13_+_23104134 | 0.78 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr8_+_15277874 | 0.78 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr25_+_29688671 | 0.78 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr3_-_34561624 | 0.78 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr11_+_44502410 | 0.78 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr7_+_24814866 | 0.77 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr23_-_28423222 | 0.77 |
ENSDART00000182058
|
c1ql4a
|
complement component 1, q subcomponent-like 4 |
| chr6_+_18418651 | 0.77 |
ENSDART00000171097
ENSDART00000167798 |
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr24_-_5691956 | 0.77 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr1_+_12394205 | 0.76 |
ENSDART00000138622
ENSDART00000136421 ENSDART00000139440 ENSDART00000184296 ENSDART00000008127 |
zgc:77739
|
zgc:77739 |
| chr23_-_14769523 | 0.76 |
ENSDART00000054909
|
gss
|
glutathione synthetase |
| chr8_-_16725959 | 0.76 |
ENSDART00000183593
|
depdc1a
|
DEP domain containing 1a |
| chr16_-_10261652 | 0.75 |
ENSDART00000163599
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr16_+_38820486 | 0.75 |
ENSDART00000131866
|
trhra
|
thyrotropin-releasing hormone receptor a |
| chr24_-_24271629 | 0.74 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr16_+_38820660 | 0.73 |
ENSDART00000182538
ENSDART00000189879 |
trhra
|
thyrotropin-releasing hormone receptor a |
| chr12_+_30368145 | 0.73 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr20_+_13141408 | 0.73 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr1_+_15258641 | 0.72 |
ENSDART00000033018
|
pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr18_+_14684115 | 0.71 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr11_+_2458264 | 0.71 |
ENSDART00000188341
|
tp53rk
|
TP53 regulating kinase |
| chr13_+_49727333 | 0.71 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr19_+_11432330 | 0.71 |
ENSDART00000188065
|
PQLC1
|
PQ loop repeat containing 1 |
| chr7_+_13684012 | 0.69 |
ENSDART00000056893
|
pdcd7
|
programmed cell death 7 |
| chr6_-_15065376 | 0.69 |
ENSDART00000087797
|
tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr23_-_28423048 | 0.69 |
ENSDART00000140448
|
c1ql4a
|
complement component 1, q subcomponent-like 4 |
| chr7_-_16597130 | 0.68 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr21_+_34981263 | 0.68 |
ENSDART00000132711
|
rbm11
|
RNA binding motif protein 11 |
| chr1_+_12177195 | 0.68 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr11_+_25157374 | 0.68 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr13_+_15580758 | 0.68 |
ENSDART00000087194
ENSDART00000013525 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr23_-_19684721 | 0.67 |
ENSDART00000184325
|
zgc:193598
|
zgc:193598 |
| chr7_-_16596938 | 0.67 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr25_+_7532811 | 0.67 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr2_-_31800521 | 0.66 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr5_-_69482891 | 0.66 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr19_-_3821678 | 0.66 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr4_-_3353595 | 0.65 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr8_+_19977712 | 0.64 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr22_-_10539180 | 0.63 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr3_+_46315016 | 0.63 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr13_+_7665890 | 0.63 |
ENSDART00000046792
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr8_-_13735572 | 0.63 |
ENSDART00000139642
|
si:dkey-258f14.7
|
si:dkey-258f14.7 |
| chr10_+_29771256 | 0.63 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr19_-_10915898 | 0.63 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr2_+_45696743 | 0.63 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr22_-_31020690 | 0.63 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr21_+_3941758 | 0.62 |
ENSDART00000181345
|
setx
|
senataxin |
| chr6_+_13046720 | 0.61 |
ENSDART00000165896
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr25_+_28693451 | 0.61 |
ENSDART00000148366
|
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr11_+_25560632 | 0.61 |
ENSDART00000033914
|
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr3_-_36364903 | 0.61 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr23_-_4225830 | 0.61 |
ENSDART00000170455
|
aar2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr15_+_25489406 | 0.61 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr2_-_32738535 | 0.60 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr11_+_6882362 | 0.60 |
ENSDART00000144181
|
klhl26
|
kelch-like family member 26 |
| chr7_+_26545502 | 0.60 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr16_-_17586883 | 0.60 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr17_-_28811747 | 0.60 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr8_-_20862443 | 0.60 |
ENSDART00000147267
|
si:ch211-133l5.8
|
si:ch211-133l5.8 |
| chr1_-_34685329 | 0.60 |
ENSDART00000125944
ENSDART00000008277 |
pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr17_+_45259259 | 0.60 |
ENSDART00000109532
ENSDART00000154890 |
ganc
|
glucosidase, alpha; neutral C |
| chr18_+_3243292 | 0.59 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr5_+_23913585 | 0.59 |
ENSDART00000015401
|
ercc6l
|
excision repair cross-complementation group 6-like |
| chr3_-_55511569 | 0.59 |
ENSDART00000108995
|
tex2
|
testis expressed 2 |
| chr6_-_40713183 | 0.59 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr8_-_32385989 | 0.59 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr8_-_35960987 | 0.59 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr5_+_39099172 | 0.58 |
ENSDART00000006079
|
bmp2k
|
BMP2 inducible kinase |
| chr22_+_35275468 | 0.58 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr9_-_30385059 | 0.58 |
ENSDART00000060134
|
piga
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr8_-_30424182 | 0.58 |
ENSDART00000099021
|
dock8
|
dedicator of cytokinesis 8 |
| chr12_+_47794089 | 0.58 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr7_+_24573721 | 0.58 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr12_-_9790485 | 0.58 |
ENSDART00000027321
|
prdm9
|
PR domain containing 9 |
| chr19_-_2085027 | 0.57 |
ENSDART00000063615
|
snx13
|
sorting nexin 13 |
| chr16_+_33992418 | 0.57 |
ENSDART00000101885
ENSDART00000130540 |
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr5_-_22969424 | 0.57 |
ENSDART00000143869
ENSDART00000172549 |
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr9_-_32648725 | 0.57 |
ENSDART00000123374
|
dnajb11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr7_-_50764714 | 0.57 |
ENSDART00000110283
|
iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr2_+_35603637 | 0.57 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr3_-_16719244 | 0.57 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr19_-_31765615 | 0.56 |
ENSDART00000103636
|
si:dkeyp-120h9.1
|
si:dkeyp-120h9.1 |
| chr9_-_704667 | 0.56 |
ENSDART00000147092
|
cflarb
|
CASP8 and FADD-like apoptosis regulator b |
| chr5_-_57723929 | 0.56 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr5_+_39099380 | 0.56 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr17_-_29213710 | 0.56 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr19_+_348729 | 0.56 |
ENSDART00000114284
|
mcl1a
|
MCL1, BCL2 family apoptosis regulator a |
| chr14_-_14687004 | 0.56 |
ENSDART00000169970
|
gcna
|
germ cell nuclear acidic peptidase |
| chr8_+_25959940 | 0.56 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr1_+_44554105 | 0.55 |
ENSDART00000149764
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr3_+_47131112 | 0.55 |
ENSDART00000165100
|
dnm2a
|
dynamin 2a |
| chr5_-_25733745 | 0.55 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr14_+_41406321 | 0.55 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr9_+_13638329 | 0.55 |
ENSDART00000143432
|
als2a
|
amyotrophic lateral sclerosis 2a (juvenile) |
| chr5_-_24029228 | 0.55 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr2_+_7557912 | 0.55 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr25_-_24240797 | 0.54 |
ENSDART00000132790
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr12_-_35944654 | 0.54 |
ENSDART00000162579
ENSDART00000164199 |
dnai2a
|
dynein, axonemal, intermediate chain 2a |
| chr23_+_7710447 | 0.54 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr22_+_35275206 | 0.54 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr17_-_25861787 | 0.54 |
ENSDART00000182503
|
BX000364.5
|
|
| chr7_+_26545911 | 0.54 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr21_-_7781555 | 0.54 |
ENSDART00000084380
ENSDART00000189131 |
aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr20_+_27087539 | 0.54 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr11_-_36009924 | 0.54 |
ENSDART00000189959
ENSDART00000167472 ENSDART00000191211 ENSDART00000191662 ENSDART00000191780 ENSDART00000192622 ENSDART00000179911 |
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr4_-_77252368 | 0.53 |
ENSDART00000111941
|
zgc:174310
|
zgc:174310 |
| chr6_+_49723289 | 0.53 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr20_-_18794789 | 0.53 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr19_+_31873308 | 0.53 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr9_+_70157 | 0.53 |
ENSDART00000140961
ENSDART00000192079 ENSDART00000157154 |
RANBP2
|
si:ch1073-55a19.2 |
| chr9_-_27719998 | 0.53 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr3_+_46479913 | 0.53 |
ENSDART00000149755
|
tyk2
|
tyrosine kinase 2 |
| chr1_-_47114310 | 0.52 |
ENSDART00000144899
ENSDART00000053157 |
setd4
|
SET domain containing 4 |
| chr7_+_57088920 | 0.52 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr7_+_32901658 | 0.52 |
ENSDART00000115420
|
ano9b
|
anoctamin 9b |
| chr7_+_41887429 | 0.52 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr7_+_25015151 | 0.52 |
ENSDART00000149966
ENSDART00000175583 |
b3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr9_+_24088062 | 0.52 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr5_+_27137473 | 0.52 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr1_+_494297 | 0.52 |
ENSDART00000108579
ENSDART00000146732 |
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr16_+_26439518 | 0.52 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr15_-_25365319 | 0.52 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr4_-_9867476 | 0.52 |
ENSDART00000091630
|
fbxo18
|
F-box protein, helicase, 18 |
| chr19_+_42694211 | 0.51 |
ENSDART00000180153
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr6_+_29861288 | 0.51 |
ENSDART00000166782
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_40232615 | 0.51 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr17_-_24587686 | 0.51 |
ENSDART00000143084
|
aftphb
|
aftiphilin b |
| chr24_-_23839647 | 0.51 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr21_-_31207522 | 0.50 |
ENSDART00000191637
|
zgc:152891
|
zgc:152891 |
| chr12_-_17810543 | 0.50 |
ENSDART00000090484
|
tecpr1a
|
tectonin beta-propeller repeat containing 1a |
| chr23_+_17980875 | 0.50 |
ENSDART00000163452
|
chia.6
|
chitinase, acidic.6 |
| chr19_+_43828104 | 0.50 |
ENSDART00000166031
|
eloa
|
elongin A |
| chr9_+_32649088 | 0.50 |
ENSDART00000136136
ENSDART00000172033 |
rabl3
|
RAB, member of RAS oncogene family-like 3 |
| chr7_-_40578733 | 0.50 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr19_-_3741602 | 0.50 |
ENSDART00000170301
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr23_-_36823932 | 0.50 |
ENSDART00000142305
|
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr3_-_35865040 | 0.49 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr20_-_34711528 | 0.49 |
ENSDART00000061555
|
si:ch211-63o20.7
|
si:ch211-63o20.7 |
| chr23_-_24955135 | 0.49 |
ENSDART00000136347
ENSDART00000144903 |
zbtb48
|
zinc finger and BTB domain containing 48 |
| chr24_-_31092711 | 0.49 |
ENSDART00000168398
|
hccsb
|
holocytochrome c synthase b |
| chr19_+_4066449 | 0.49 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr5_-_41124241 | 0.49 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr24_-_24796583 | 0.49 |
ENSDART00000144791
ENSDART00000146570 |
pde7a
|
phosphodiesterase 7A |
| chr10_+_26990095 | 0.49 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr14_-_45558490 | 0.49 |
ENSDART00000165060
|
ints5
|
integrator complex subunit 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of spi2+spic
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.4 | 1.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 1.4 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.3 | 1.0 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.3 | 1.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.9 | GO:1904590 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.2 | 0.6 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.2 | 0.6 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.2 | 0.6 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.2 | 0.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 0.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 0.9 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 1.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 0.5 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 0.7 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 0.8 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 1.0 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 0.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 0.7 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.2 | 0.6 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.2 | 0.5 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 0.5 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.2 | 0.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 2.0 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.6 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.1 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.7 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.9 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.5 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.6 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 1.9 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.3 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.8 | GO:0072425 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.5 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 0.5 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.4 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 0.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.4 | GO:0071265 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.0 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.6 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 1.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.4 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.7 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.8 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 1.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.5 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.9 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.1 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.4 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.1 | 0.3 | GO:1904867 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.3 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.2 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.1 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.9 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.4 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 1.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.5 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.4 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 1.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.7 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.9 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 1.2 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.5 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.2 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0055021 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.5 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.7 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.4 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 1.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.5 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 1.8 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.4 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.1 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.8 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 1.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 1.5 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:0042531 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.2 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.3 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.5 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 2.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.1 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.9 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.0 | 0.3 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.6 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.2 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.8 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0016575 | histone deacetylation(GO:0016575) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.9 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.2 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 1.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 2.0 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 5.7 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.9 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.5 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 1.5 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.2 | 0.6 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.2 | 0.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 1.0 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 1.4 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 1.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.2 | 0.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.6 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 1.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 1.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.4 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.3 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.7 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.5 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 0.4 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 2.1 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.5 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.6 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.6 | GO:0052813 | phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.6 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.9 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 1.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.7 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 1.8 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 1.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.3 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 4.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 2.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.5 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.0 | 0.3 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |