Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
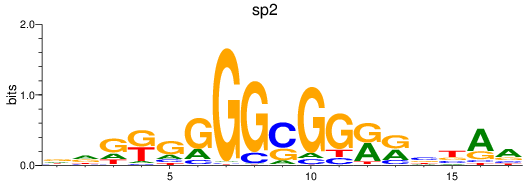
Results for sp2
Z-value: 2.04
Transcription factors associated with sp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp2
|
ENSDARG00000076763 | sp2 transcription factor |
|
sp2
|
ENSDARG00000113443 | sp2 transcription factor |
|
sp2
|
ENSDARG00000115477 | sp2 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp2 | dr11_v1_chr11_-_12008001_12008001 | 0.91 | 7.0e-04 | Click! |
Activity profile of sp2 motif
Sorted Z-values of sp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33991801 | 5.55 |
ENSDART00000061744
|
zp3a.1
|
zona pellucida glycoprotein 3a, tandem duplicate 1 |
| chr17_-_2584423 | 5.04 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr20_+_33987465 | 4.94 |
ENSDART00000061751
|
zp3a.2
|
zona pellucida glycoprotein 3a, tandem duplicate 2 |
| chr24_-_20599781 | 4.76 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr22_+_835728 | 4.05 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr18_-_127558 | 3.47 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr20_+_54304800 | 3.38 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr13_-_17860307 | 3.03 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr25_+_22319940 | 2.97 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr10_-_641609 | 2.89 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr12_-_23365737 | 2.88 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr19_+_26340736 | 2.78 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr23_+_2740741 | 2.71 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr12_+_46634736 | 2.69 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr3_+_29469283 | 2.58 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr20_+_39250673 | 2.58 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr10_+_1849874 | 2.52 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr2_+_59015878 | 2.40 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr18_-_127873 | 2.31 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr8_-_4100365 | 2.31 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr18_-_41161828 | 2.27 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr7_-_45852270 | 2.27 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr3_+_35611625 | 2.25 |
ENSDART00000190995
|
traf7
|
TNF receptor-associated factor 7 |
| chr3_+_51684963 | 2.16 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr15_+_25489406 | 2.16 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr14_-_237130 | 2.14 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr3_-_16227683 | 2.13 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr5_-_30074332 | 2.13 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr3_+_46559639 | 2.13 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr16_+_25259313 | 2.12 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr23_+_2825940 | 2.12 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr24_-_26622423 | 2.10 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr16_-_7793457 | 2.09 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr5_-_12743196 | 2.09 |
ENSDART00000188976
ENSDART00000137705 |
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr23_+_30898013 | 2.09 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr20_-_182841 | 2.09 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr13_-_12021566 | 2.05 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr7_-_32020100 | 2.04 |
ENSDART00000185433
|
kif18a
|
kinesin family member 18A |
| chr24_-_36593876 | 2.00 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr5_-_30080332 | 1.95 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr18_+_27077853 | 1.92 |
ENSDART00000125326
ENSDART00000192660 ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr6_+_475264 | 1.88 |
ENSDART00000193615
|
LO017974.1
|
|
| chr15_+_29393519 | 1.84 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr20_-_1268863 | 1.83 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr21_-_40557281 | 1.83 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr20_-_14781904 | 1.78 |
ENSDART00000187200
ENSDART00000179912 ENSDART00000160481 ENSDART00000026969 |
suco
|
SUN domain containing ossification factor |
| chr1_-_45616242 | 1.77 |
ENSDART00000150066
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_+_1473929 | 1.76 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr7_-_48396193 | 1.71 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr1_-_45616470 | 1.69 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr21_-_43398457 | 1.68 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr20_+_46741074 | 1.65 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr11_-_35975026 | 1.62 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr8_+_23165749 | 1.58 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr10_+_39199547 | 1.57 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr21_+_21279159 | 1.57 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr16_-_2870522 | 1.57 |
ENSDART00000148543
|
cdcp1a
|
CUB domain containing protein 1a |
| chr2_+_25657958 | 1.54 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr20_-_16156419 | 1.53 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr15_-_9421481 | 1.52 |
ENSDART00000189045
ENSDART00000177158 |
sacs
|
sacsin molecular chaperone |
| chr22_+_31025096 | 1.51 |
ENSDART00000185953
|
zmp:0000000735
|
zmp:0000000735 |
| chr8_+_2656231 | 1.51 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr2_+_25658112 | 1.51 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr7_+_38962207 | 1.50 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr19_+_13994563 | 1.49 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr16_-_1757521 | 1.47 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr13_+_23093743 | 1.47 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr6_+_41503854 | 1.46 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr22_+_883678 | 1.44 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr21_-_11054605 | 1.43 |
ENSDART00000191378
ENSDART00000084061 |
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr13_+_16521898 | 1.43 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr22_-_16275236 | 1.41 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr24_-_28381404 | 1.40 |
ENSDART00000148406
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr10_+_15603082 | 1.40 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr11_+_19370717 | 1.38 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr19_-_26769867 | 1.37 |
ENSDART00000043776
ENSDART00000159489 ENSDART00000138675 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr8_-_5847533 | 1.37 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr2_-_58075414 | 1.37 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr15_-_25094026 | 1.35 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr16_-_5844881 | 1.34 |
ENSDART00000085678
|
trak1a
|
trafficking protein, kinesin binding 1a |
| chr21_-_43398122 | 1.34 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr18_-_26715156 | 1.33 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr9_+_55536005 | 1.33 |
ENSDART00000192364
|
mxra5b
|
matrix-remodelling associated 5b |
| chr3_+_35812040 | 1.33 |
ENSDART00000075903
ENSDART00000147712 |
crlf3
|
cytokine receptor-like factor 3 |
| chr3_+_16841942 | 1.32 |
ENSDART00000023985
ENSDART00000145317 |
stk17al
|
serine/threonine kinase 17a like |
| chr6_+_3334392 | 1.32 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr25_+_3104959 | 1.32 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr24_+_10039165 | 1.30 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr15_-_25209308 | 1.30 |
ENSDART00000157857
|
mnta
|
MAX network transcriptional repressor a |
| chr8_+_8166285 | 1.29 |
ENSDART00000147940
|
plxnb3
|
plexin B3 |
| chr17_-_40956035 | 1.27 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr21_+_43253538 | 1.27 |
ENSDART00000179940
ENSDART00000164806 ENSDART00000147026 |
shroom1
|
shroom family member 1 |
| chr12_-_9132682 | 1.25 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr21_-_11054876 | 1.25 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr15_-_25093680 | 1.24 |
ENSDART00000062695
|
exo5
|
exonuclease 5 |
| chr2_-_24061575 | 1.24 |
ENSDART00000089234
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr11_-_16395956 | 1.21 |
ENSDART00000115085
|
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr11_+_19370447 | 1.21 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr5_+_61361815 | 1.21 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr8_-_22739757 | 1.19 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr19_+_4892281 | 1.18 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr3_-_16227490 | 1.14 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr10_+_39200213 | 1.12 |
ENSDART00000153727
|
ei24
|
etoposide induced 2.4 |
| chr19_-_46088429 | 1.11 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr19_-_26770083 | 1.11 |
ENSDART00000193811
ENSDART00000174455 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr7_+_55112922 | 1.10 |
ENSDART00000073549
|
snai3
|
snail family zinc finger 3 |
| chr18_-_2727764 | 1.10 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr12_+_23850661 | 1.10 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr9_+_23825440 | 1.08 |
ENSDART00000138470
|
ints6
|
integrator complex subunit 6 |
| chr11_-_44030962 | 1.08 |
ENSDART00000171910
|
FP016005.1
|
|
| chr15_+_47618221 | 1.07 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr25_+_16356083 | 1.07 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr22_+_28236737 | 1.06 |
ENSDART00000086868
|
senp7b
|
SUMO1/sentrin specific peptidase 7b |
| chr20_+_9474841 | 1.06 |
ENSDART00000053847
|
rad51b
|
RAD51 paralog B |
| chr16_+_7991274 | 1.05 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr8_+_39802506 | 1.05 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr19_+_935565 | 1.02 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr24_-_10828560 | 1.01 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr9_-_53899106 | 1.01 |
ENSDART00000171986
|
cog3
|
component of oligomeric golgi complex 3 |
| chr25_-_6389713 | 1.01 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr10_+_37500234 | 0.99 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr13_-_26799244 | 0.99 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr19_+_390298 | 0.98 |
ENSDART00000136361
|
sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr17_+_44756247 | 0.97 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr6_-_60104628 | 0.96 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr14_+_30910114 | 0.95 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr2_+_44666705 | 0.95 |
ENSDART00000139137
|
yeats2
|
YEATS domain containing 2 |
| chr20_+_812012 | 0.94 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr20_+_36820965 | 0.94 |
ENSDART00000153085
ENSDART00000062935 |
heca
|
hdc homolog, cell cycle regulator |
| chr14_+_28545198 | 0.94 |
ENSDART00000125362
ENSDART00000105902 |
hmmr
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr25_+_3507368 | 0.94 |
ENSDART00000157777
|
zgc:153293
|
zgc:153293 |
| chr1_-_55210619 | 0.92 |
ENSDART00000111671
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr13_-_18069421 | 0.91 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr2_+_105748 | 0.91 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr22_+_12366516 | 0.90 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr5_+_72152813 | 0.90 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr11_-_40504170 | 0.88 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr2_+_16696052 | 0.88 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr12_-_17492852 | 0.87 |
ENSDART00000012421
ENSDART00000138766 ENSDART00000130735 |
minpp1b
|
multiple inositol-polyphosphate phosphatase 1b |
| chr20_-_28931901 | 0.87 |
ENSDART00000153082
ENSDART00000046042 |
susd6
|
sushi domain containing 6 |
| chr2_+_24867534 | 0.86 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr9_-_704667 | 0.86 |
ENSDART00000147092
|
cflarb
|
CASP8 and FADD-like apoptosis regulator b |
| chr3_-_33941875 | 0.85 |
ENSDART00000047660
|
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr15_+_3125136 | 0.84 |
ENSDART00000130968
|
rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr6_+_3334710 | 0.82 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr14_-_15154695 | 0.82 |
ENSDART00000160677
|
uvssa
|
UV-stimulated scaffold protein A |
| chr19_-_31900893 | 0.82 |
ENSDART00000113797
|
zbtb10
|
zinc finger and BTB domain containing 10 |
| chr15_-_43768776 | 0.80 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr14_-_46113321 | 0.80 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr16_-_17586883 | 0.80 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr1_-_59232267 | 0.80 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr3_+_14499075 | 0.80 |
ENSDART00000162853
ENSDART00000167776 ENSDART00000162932 ENSDART00000168229 |
tmem205
|
transmembrane protein 205 |
| chr21_-_13972745 | 0.79 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr7_+_38962459 | 0.79 |
ENSDART00000173851
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr17_+_51746830 | 0.79 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr22_-_5918670 | 0.78 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr23_-_30960506 | 0.78 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr17_+_21546993 | 0.77 |
ENSDART00000182387
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr19_+_1097393 | 0.77 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr10_-_42297889 | 0.77 |
ENSDART00000099262
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr19_+_6990970 | 0.76 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr9_-_52920305 | 0.76 |
ENSDART00000166906
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr7_+_22792132 | 0.75 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr25_+_4635355 | 0.72 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr7_-_16598212 | 0.72 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr5_-_13086616 | 0.72 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr23_+_41200854 | 0.72 |
ENSDART00000109567
|
nhsa
|
Nance-Horan syndrome a (congenital cataracts and dental anomalies) |
| chr15_-_25435085 | 0.71 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr22_-_26558166 | 0.71 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr4_-_5795309 | 0.71 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr1_-_12393060 | 0.68 |
ENSDART00000079782
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr1_-_55196103 | 0.68 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr17_-_23709347 | 0.68 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr7_-_6444011 | 0.68 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr12_-_14211293 | 0.67 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr14_+_1719367 | 0.65 |
ENSDART00000157696
|
trpc7b
|
transient receptor potential cation channel, subfamily C, member 7b |
| chr3_-_36440705 | 0.65 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr7_+_13988075 | 0.64 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr15_-_47956388 | 0.63 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr3_-_33941319 | 0.63 |
ENSDART00000026090
ENSDART00000111878 |
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr9_+_13714379 | 0.62 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr17_-_48944465 | 0.62 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr7_-_6429031 | 0.62 |
ENSDART00000173413
|
zgc:112234
|
zgc:112234 |
| chr14_-_32503363 | 0.62 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr12_+_14149686 | 0.61 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr12_-_37449396 | 0.60 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr5_+_20319519 | 0.59 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr23_-_18982499 | 0.59 |
ENSDART00000012507
|
bcl2l1
|
bcl2-like 1 |
| chr2_+_24868010 | 0.59 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr19_+_12406583 | 0.56 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr13_-_45022527 | 0.56 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr3_+_11548516 | 0.56 |
ENSDART00000059117
|
mmd
|
monocyte to macrophage differentiation-associated |
| chr24_+_42132962 | 0.56 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr6_+_2271559 | 0.55 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr19_-_2707048 | 0.54 |
ENSDART00000166112
|
il6
|
interleukin 6 (interferon, beta 2) |
| chr22_+_94692 | 0.53 |
ENSDART00000114777
|
nckipsd
|
NCK interacting protein with SH3 domain |
| chr17_+_27723490 | 0.53 |
ENSDART00000123588
ENSDART00000170462 ENSDART00000169708 |
qkia
|
QKI, KH domain containing, RNA binding a |
| chr18_-_22094102 | 0.51 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr20_-_51727860 | 0.51 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr1_+_604127 | 0.51 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.9 | 15.5 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.8 | 3.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.7 | 2.1 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.6 | 2.5 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.6 | 3.0 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.5 | 1.6 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.5 | 1.4 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.5 | 4.1 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.4 | 2.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.4 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 2.7 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.3 | 2.7 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.3 | 1.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 1.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.3 | 0.8 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.3 | 2.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.3 | 3.5 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 2.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 2.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.2 | 0.7 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 1.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 0.8 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 1.5 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 0.7 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.2 | 0.7 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.2 | 1.8 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 3.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 1.9 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.2 | 1.8 | GO:0003139 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.1 | 2.9 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 1.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.9 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 1.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.3 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 2.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.6 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.8 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 1.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 3.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.6 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 3.0 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 2.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.8 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.4 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 1.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.4 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.6 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 1.3 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.8 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 2.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.6 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.5 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 1.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.8 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 1.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 2.1 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.0 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.9 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.4 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 3.7 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 2.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.8 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 2.3 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.2 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.8 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 2.3 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 2.6 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.7 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.0 | GO:0060039 | pericardium development(GO:0060039) visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.9 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.5 | GO:0046578 | regulation of Ras protein signal transduction(GO:0046578) |
| 0.0 | 0.5 | GO:0001841 | neural tube formation(GO:0001841) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.4 | 1.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.4 | 2.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 1.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.3 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 2.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 1.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 2.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.5 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 4.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 3.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 5.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 11.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.7 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 1.0 | 3.0 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.9 | 2.6 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.5 | 6.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.5 | 4.1 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.4 | 1.5 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.3 | 2.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 2.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 0.7 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.9 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 2.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 2.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.8 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 1.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 3.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.3 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 1.6 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 3.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 2.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.7 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.8 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 2.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 3.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 2.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.2 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.6 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.7 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 3.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 6.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 9.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 2.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 3.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 4.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 1.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 3.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 2.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 2.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.9 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |