Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
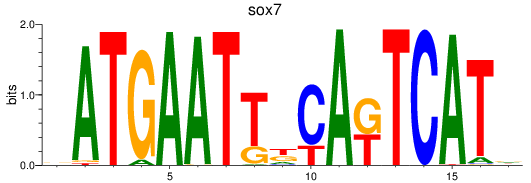
Results for sox7
Z-value: 0.37
Transcription factors associated with sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox7
|
ENSDARG00000030125 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox7 | dr11_v1_chr20_+_19066596_19066596 | -0.19 | 6.2e-01 | Click! |
Activity profile of sox7 motif
Sorted Z-values of sox7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_37103487 | 0.52 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr3_-_6709938 | 0.47 |
ENSDART00000172196
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr17_-_26537928 | 0.46 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr5_-_3960161 | 0.44 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr16_-_42306597 | 0.39 |
ENSDART00000158046
|
ppox
|
protoporphyrinogen oxidase |
| chr8_+_2787042 | 0.37 |
ENSDART00000155275
|
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr25_+_28253844 | 0.35 |
ENSDART00000151891
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr8_-_22514918 | 0.30 |
ENSDART00000021514
ENSDART00000189272 |
apex2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr18_+_15876385 | 0.29 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr7_-_4107423 | 0.29 |
ENSDART00000172971
|
zgc:55733
|
zgc:55733 |
| chr11_+_6010177 | 0.27 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr11_-_35970966 | 0.26 |
ENSDART00000188805
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr7_+_18176162 | 0.26 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr22_+_6740039 | 0.25 |
ENSDART00000144122
|
CT583625.2
|
|
| chr8_+_23174137 | 0.23 |
ENSDART00000189470
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr5_+_13891202 | 0.23 |
ENSDART00000193992
|
hk2
|
hexokinase 2 |
| chr18_+_14684115 | 0.19 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr15_-_21692630 | 0.19 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr11_-_34577034 | 0.19 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr14_-_6365223 | 0.19 |
ENSDART00000167446
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr18_-_12654011 | 0.19 |
ENSDART00000062343
|
hipk2
|
homeodomain interacting protein kinase 2 |
| chr12_-_33558879 | 0.18 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr8_-_21058354 | 0.17 |
ENSDART00000130481
ENSDART00000113749 |
zgc:112962
|
zgc:112962 |
| chr22_-_26558166 | 0.16 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr2_+_27386617 | 0.16 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr21_+_20903244 | 0.16 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr20_+_37294112 | 0.15 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr14_+_1124409 | 0.15 |
ENSDART00000190043
ENSDART00000106708 ENSDART00000160246 |
sec24b
|
SEC24 homolog B, COPII coat complex component |
| chr21_+_19330774 | 0.15 |
ENSDART00000109412
|
helq
|
helicase, POLQ like |
| chr22_-_12862415 | 0.15 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr11_+_14104417 | 0.14 |
ENSDART00000059752
ENSDART00000186575 |
med16
|
mediator complex subunit 16 |
| chr18_-_18524624 | 0.14 |
ENSDART00000157834
|
lonp2
|
lon peptidase 2, peroxisomal |
| chr8_-_40205712 | 0.14 |
ENSDART00000158927
|
anapc5
|
anaphase promoting complex subunit 5 |
| chr1_+_6206950 | 0.13 |
ENSDART00000146473
|
retreg2
|
reticulophagy regulator family member 2 |
| chr14_-_30905963 | 0.13 |
ENSDART00000183543
ENSDART00000186441 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr10_+_29770120 | 0.13 |
ENSDART00000100032
ENSDART00000193205 |
hyou1
|
hypoxia up-regulated 1 |
| chr22_-_26852516 | 0.12 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr7_+_18878973 | 0.12 |
ENSDART00000037846
|
focad
|
focadhesin |
| chr8_-_16650595 | 0.12 |
ENSDART00000135319
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr18_-_39200557 | 0.12 |
ENSDART00000132367
ENSDART00000183672 |
si:ch211-235f12.2
mapk6
|
si:ch211-235f12.2 mitogen-activated protein kinase 6 |
| chr11_-_3308569 | 0.12 |
ENSDART00000036581
|
cdk2
|
cyclin-dependent kinase 2 |
| chr16_+_27345383 | 0.12 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr14_-_30490465 | 0.11 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr21_+_4320769 | 0.11 |
ENSDART00000168553
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr13_-_30161684 | 0.10 |
ENSDART00000040409
|
ppa1b
|
pyrophosphatase (inorganic) 1b |
| chr11_+_44356707 | 0.09 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr20_-_4031475 | 0.09 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr19_-_3783141 | 0.08 |
ENSDART00000162260
|
btr19
|
bloodthirsty-related gene family, member 19 |
| chr12_+_30109698 | 0.08 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr23_+_21261313 | 0.08 |
ENSDART00000104268
ENSDART00000159046 |
emc1
|
ER membrane protein complex subunit 1 |
| chr25_-_7999756 | 0.08 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr10_-_7913591 | 0.07 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr3_-_27065477 | 0.07 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr12_-_4540564 | 0.07 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr15_+_15771418 | 0.07 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr11_+_44356504 | 0.06 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr21_-_18932761 | 0.06 |
ENSDART00000140129
|
med15
|
mediator complex subunit 15 |
| chr20_-_48898371 | 0.06 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr24_-_25166416 | 0.06 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr10_+_27068251 | 0.06 |
ENSDART00000012717
|
ehd1b
|
EH-domain containing 1b |
| chr20_-_48898560 | 0.05 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr11_-_3613558 | 0.05 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr20_+_50956369 | 0.05 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr7_-_13648635 | 0.05 |
ENSDART00000148957
ENSDART00000130967 |
spg21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr19_+_5480327 | 0.05 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr3_+_52737565 | 0.04 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr13_+_43650632 | 0.03 |
ENSDART00000141024
|
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr21_-_43171249 | 0.03 |
ENSDART00000021037
ENSDART00000148630 |
hspa4a
|
heat shock protein 4a |
| chr6_+_49081141 | 0.03 |
ENSDART00000131080
ENSDART00000050000 |
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr20_-_42534153 | 0.03 |
ENSDART00000061122
|
rfx6
|
regulatory factor X, 6 |
| chr21_-_19330333 | 0.02 |
ENSDART00000101462
|
mrps18c
|
mitochondrial ribosomal protein S18C |
| chr22_+_31330485 | 0.02 |
ENSDART00000192585
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr2_-_45504798 | 0.02 |
ENSDART00000131412
|
gpsm2
|
G protein signaling modulator 2 |
| chr10_-_4961923 | 0.02 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr1_+_12335816 | 0.01 |
ENSDART00000067086
|
nansa
|
N-acetylneuraminic acid synthase a |
| chr9_+_38369872 | 0.00 |
ENSDART00000193914
|
plcd4b
|
phospholipase C, delta 4b |
| chr8_+_16726386 | 0.00 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr7_-_35083184 | 0.00 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr8_-_46457233 | 0.00 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.4 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.2 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.0 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |