Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
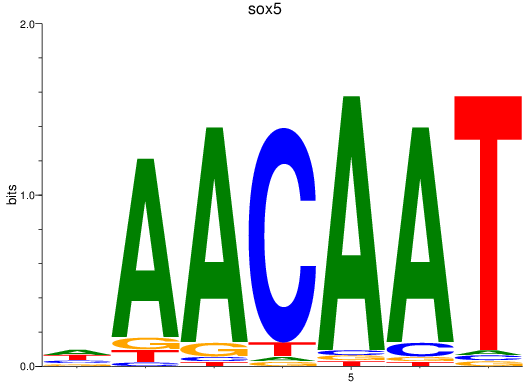
Results for sox5
Z-value: 2.06
Transcription factors associated with sox5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox5
|
ENSDARG00000011582 | SRY-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox5 | dr11_v1_chr4_-_17055950_17055950 | 0.96 | 3.6e-05 | Click! |
Activity profile of sox5 motif
Sorted Z-values of sox5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22339582 | 5.87 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr14_+_38786298 | 4.15 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr8_+_21195420 | 3.98 |
ENSDART00000100234
ENSDART00000091307 |
col2a1a
|
collagen, type II, alpha 1a |
| chr7_+_39444843 | 3.32 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr9_-_22135420 | 3.25 |
ENSDART00000184959
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr15_+_28685892 | 3.13 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr9_-_22272181 | 3.06 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr18_+_16246806 | 3.02 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr16_+_10318893 | 2.95 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr5_+_35458190 | 2.93 |
ENSDART00000051313
|
fbp1b
|
fructose-1,6-bisphosphatase 1b |
| chr24_+_24461558 | 2.79 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr24_+_24461341 | 2.77 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr11_-_6048490 | 2.70 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr3_-_31804481 | 2.68 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr25_+_14507567 | 2.67 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr19_+_19777437 | 2.56 |
ENSDART00000170662
|
hoxa3a
|
homeobox A3a |
| chr14_-_25949951 | 2.53 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr20_+_10538025 | 2.46 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr10_+_36650222 | 2.46 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr20_-_14054083 | 2.44 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr21_-_25741411 | 2.44 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr4_-_16412084 | 2.40 |
ENSDART00000188460
|
dcn
|
decorin |
| chr22_-_15602760 | 2.36 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr3_-_35602233 | 2.34 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr20_-_35246150 | 2.32 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr19_+_19759577 | 2.23 |
ENSDART00000169480
|
hoxa5a
|
homeobox A5a |
| chr21_-_35853245 | 2.21 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr9_+_32978302 | 2.19 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr20_-_47704973 | 2.18 |
ENSDART00000174808
|
tfap2b
|
transcription factor AP-2 beta |
| chr5_-_28606916 | 2.15 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr14_-_24761132 | 2.12 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr10_+_31244619 | 2.12 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr14_-_25599002 | 1.99 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr22_-_15602593 | 1.99 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr20_+_34915945 | 1.96 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr5_-_14344647 | 1.95 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr19_+_19762183 | 1.91 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr16_-_28856112 | 1.91 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr13_+_25449681 | 1.90 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr8_+_23093155 | 1.89 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr1_+_53945934 | 1.83 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr8_-_9118958 | 1.82 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr23_+_8797143 | 1.81 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr7_-_51102479 | 1.81 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr13_-_22771708 | 1.79 |
ENSDART00000112900
|
si:ch211-150i13.1
|
si:ch211-150i13.1 |
| chr1_-_38709551 | 1.79 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr2_-_50372467 | 1.78 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr3_+_23742868 | 1.77 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr3_+_23697997 | 1.74 |
ENSDART00000184299
ENSDART00000078466 |
hoxb3a
|
homeobox B3a |
| chr22_-_26945493 | 1.72 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr16_+_19732543 | 1.71 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr7_-_58729894 | 1.70 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr7_-_52558495 | 1.69 |
ENSDART00000138263
ENSDART00000009938 ENSDART00000174292 ENSDART00000174218 ENSDART00000174335 |
tcf12
|
transcription factor 12 |
| chr2_+_56463167 | 1.68 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr5_+_37837245 | 1.68 |
ENSDART00000171617
|
epd
|
ependymin |
| chr19_-_2318391 | 1.68 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr10_+_4095917 | 1.67 |
ENSDART00000114533
|
mn1a
|
meningioma 1a |
| chr8_-_31938364 | 1.66 |
ENSDART00000137647
|
RNF180
|
si:dkey-250l23.4 |
| chr13_+_28854438 | 1.65 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr23_+_36083529 | 1.65 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr3_-_30158395 | 1.63 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr16_-_7443388 | 1.62 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr6_+_43426599 | 1.60 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr13_-_40411908 | 1.60 |
ENSDART00000057094
ENSDART00000150091 |
nkx2.3
|
NK2 homeobox 3 |
| chr23_+_36118738 | 1.60 |
ENSDART00000139319
|
hoxc5a
|
homeobox C5a |
| chr17_-_32865788 | 1.57 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr16_+_28754403 | 1.57 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr18_-_14836600 | 1.55 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr10_-_28761454 | 1.55 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr13_+_29773153 | 1.54 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr6_+_42819337 | 1.53 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr15_+_19797918 | 1.53 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr12_+_45677293 | 1.51 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr21_+_28502340 | 1.51 |
ENSDART00000077897
ENSDART00000140229 |
otub1a
|
OTU deubiquitinase, ubiquitin aldehyde binding 1a |
| chr12_-_35054354 | 1.51 |
ENSDART00000075351
|
zgc:112285
|
zgc:112285 |
| chr18_+_48428713 | 1.51 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr8_+_7311478 | 1.50 |
ENSDART00000133358
|
ssuh2rs1
|
ssu-2 homolog, related sequence 1 |
| chr8_+_13106760 | 1.50 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr9_+_22003942 | 1.49 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr13_+_29771463 | 1.47 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr5_-_67911111 | 1.46 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr11_+_25257022 | 1.44 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr17_+_29276544 | 1.43 |
ENSDART00000049399
|
ankrd9
|
ankyrin repeat domain 9 |
| chr11_-_23458792 | 1.41 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr22_+_661711 | 1.41 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr6_+_42818963 | 1.38 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr12_-_20373058 | 1.37 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr18_+_6039141 | 1.36 |
ENSDART00000138972
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr14_-_26704829 | 1.33 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr13_+_12045475 | 1.31 |
ENSDART00000163053
ENSDART00000160812 ENSDART00000158244 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr22_-_10487490 | 1.29 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr23_-_5685023 | 1.28 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr10_+_10351685 | 1.26 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr11_+_21910343 | 1.25 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr23_-_31428763 | 1.25 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr2_-_10338759 | 1.25 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr19_+_1964005 | 1.22 |
ENSDART00000172049
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr18_-_14836862 | 1.21 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr13_+_51579851 | 1.21 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr16_-_43025885 | 1.20 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr9_+_24008879 | 1.20 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr10_+_21807497 | 1.20 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr7_+_47287307 | 1.18 |
ENSDART00000114669
|
dpy19l3
|
dpy-19 like C-mannosyltransferase 3 |
| chr5_+_17624463 | 1.18 |
ENSDART00000183869
ENSDART00000081064 |
fbrsl1
|
fibrosin-like 1 |
| chr24_-_6647275 | 1.17 |
ENSDART00000161494
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr11_+_21910752 | 1.16 |
ENSDART00000114288
|
foxp4
|
forkhead box P4 |
| chr16_+_23087326 | 1.16 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr12_-_1951233 | 1.16 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr10_+_22003750 | 1.16 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr9_-_42873700 | 1.15 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr7_-_38861741 | 1.15 |
ENSDART00000173629
ENSDART00000037361 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr16_+_29492937 | 1.14 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr5_+_61998641 | 1.13 |
ENSDART00000137110
|
selenow1
|
selenoprotein W, 1 |
| chr13_+_12045758 | 1.13 |
ENSDART00000079398
ENSDART00000165467 ENSDART00000165880 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr16_+_23811554 | 1.12 |
ENSDART00000114336
|
si:dkey-7f3.9
|
si:dkey-7f3.9 |
| chr2_-_30668580 | 1.12 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr18_+_40355408 | 1.11 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr8_+_31941650 | 1.08 |
ENSDART00000138217
|
htr1aa
|
5-hydroxytryptamine (serotonin) receptor 1A a |
| chr19_-_45960191 | 1.08 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr1_-_30689004 | 1.08 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr17_-_24680965 | 1.08 |
ENSDART00000154880
|
arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr14_-_30808174 | 1.07 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr23_-_30787932 | 1.07 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr1_-_16665044 | 1.06 |
ENSDART00000040434
|
asah1b
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1b |
| chr25_-_29134654 | 1.05 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr15_-_20233105 | 1.05 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr13_-_31647323 | 1.04 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr1_-_22726233 | 1.04 |
ENSDART00000140920
|
prom1b
|
prominin 1 b |
| chr6_-_21873266 | 1.04 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr25_-_18125769 | 1.02 |
ENSDART00000140484
|
kitlga
|
kit ligand a |
| chr13_-_13294847 | 1.01 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr7_+_32369463 | 0.99 |
ENSDART00000180544
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr8_+_8291492 | 0.99 |
ENSDART00000151314
|
srpk3
|
SRSF protein kinase 3 |
| chr8_-_34051548 | 0.98 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr7_+_30970045 | 0.98 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr2_+_20539402 | 0.98 |
ENSDART00000129585
|
si:ch73-14h1.2
|
si:ch73-14h1.2 |
| chr19_-_103289 | 0.98 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr18_-_1185772 | 0.98 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr3_+_32689707 | 0.97 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr13_-_7575216 | 0.96 |
ENSDART00000159443
|
pitx3
|
paired-like homeodomain 3 |
| chr16_+_14588141 | 0.95 |
ENSDART00000140469
ENSDART00000059984 ENSDART00000167411 ENSDART00000133566 |
deptor
|
DEP domain containing MTOR-interacting protein |
| chr3_-_26204867 | 0.94 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr21_-_20765338 | 0.93 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr6_-_32093830 | 0.90 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr11_+_27274355 | 0.90 |
ENSDART00000113707
|
fbln2
|
fibulin 2 |
| chr15_+_24644016 | 0.90 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr4_-_6567355 | 0.90 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr3_-_28828242 | 0.90 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr19_+_15571290 | 0.90 |
ENSDART00000131134
|
foxo6b
|
forkhead box O6 b |
| chr15_+_24644251 | 0.89 |
ENSDART00000181660
|
smtnl
|
smoothelin, like |
| chr10_+_8968203 | 0.89 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr19_-_35596207 | 0.88 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr23_-_3681026 | 0.88 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr16_+_5678071 | 0.88 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr18_+_19972853 | 0.87 |
ENSDART00000180071
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr13_+_3667230 | 0.87 |
ENSDART00000131553
ENSDART00000189841 ENSDART00000183554 ENSDART00000018737 |
qkib
|
QKI, KH domain containing, RNA binding b |
| chr14_+_21222287 | 0.86 |
ENSDART00000159905
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr8_+_20488322 | 0.84 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr3_+_24134418 | 0.82 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr19_+_19761966 | 0.81 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr20_+_54383838 | 0.81 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr23_+_18722715 | 0.81 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr4_+_8532580 | 0.81 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr20_+_39283849 | 0.81 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr22_+_35068046 | 0.80 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr12_-_3756405 | 0.80 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr13_-_20518632 | 0.79 |
ENSDART00000165310
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr19_+_43969363 | 0.79 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr23_+_32335871 | 0.79 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr20_-_25369767 | 0.79 |
ENSDART00000180278
|
itsn2a
|
intersectin 2a |
| chr5_-_44843738 | 0.79 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr7_+_27251376 | 0.78 |
ENSDART00000173521
ENSDART00000173962 |
sox6
|
SRY (sex determining region Y)-box 6 |
| chr19_-_21832441 | 0.78 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr23_+_18722915 | 0.77 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr8_+_18830759 | 0.75 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr4_-_71904377 | 0.74 |
ENSDART00000191852
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr6_+_39812475 | 0.74 |
ENSDART00000067063
|
c1ql4b
|
complement component 1, q subcomponent-like 4b |
| chr20_+_20186706 | 0.73 |
ENSDART00000002507
|
rhoj
|
ras homolog family member J |
| chr7_-_52849913 | 0.73 |
ENSDART00000174133
ENSDART00000172951 |
map1aa
|
microtubule-associated protein 1Aa |
| chr18_-_10995410 | 0.73 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr18_+_48428126 | 0.73 |
ENSDART00000137978
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr6_-_41085443 | 0.73 |
ENSDART00000143944
ENSDART00000028217 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr9_+_22017368 | 0.73 |
ENSDART00000023059
|
zgc:153846
|
zgc:153846 |
| chr7_-_33351485 | 0.72 |
ENSDART00000146420
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr14_-_18671334 | 0.72 |
ENSDART00000182381
|
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr22_-_14128716 | 0.71 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr22_-_20342260 | 0.71 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr18_+_40354998 | 0.71 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr25_-_12923482 | 0.71 |
ENSDART00000161754
|
CR450808.1
|
|
| chr16_+_29492749 | 0.71 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr24_-_6078222 | 0.71 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr23_+_28077953 | 0.70 |
ENSDART00000186122
ENSDART00000111570 |
slc26a10
|
solute carrier family 26, member 10 |
| chr7_+_20471315 | 0.68 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr11_+_6819050 | 0.68 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr17_-_30975707 | 0.68 |
ENSDART00000138346
|
evla
|
Enah/Vasp-like a |
| chr4_-_5332814 | 0.68 |
ENSDART00000144225
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr13_+_7292061 | 0.67 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr18_+_14693682 | 0.67 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr13_-_20519001 | 0.66 |
ENSDART00000168955
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr1_+_17376922 | 0.66 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.8 | 5.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.7 | 2.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.7 | 2.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.6 | 1.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.6 | 2.4 | GO:0015840 | urea transport(GO:0015840) |
| 0.6 | 3.0 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.5 | 2.0 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.4 | 1.7 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.4 | 1.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 2.0 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.4 | 3.0 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.4 | 1.8 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.4 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 1.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.3 | 2.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 1.8 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 1.3 | GO:0048909 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.3 | 1.5 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.0 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.2 | 0.6 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.2 | 2.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 1.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 0.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 1.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 2.0 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.2 | 2.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.9 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.2 | 2.0 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 1.0 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 4.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 1.6 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 0.8 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 0.6 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 2.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 3.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 3.3 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.6 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 0.6 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 15.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 4.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.1 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 0.7 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 1.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 1.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 1.8 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.6 | GO:0089718 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 6.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.3 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 2.5 | GO:0032474 | otolith morphogenesis(GO:0032474) semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 1.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.9 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.3 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.5 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 1.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 3.3 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 1.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 2.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 2.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 2.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 6.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.8 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 0.9 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 1.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.4 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.6 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 2.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 2.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 1.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 1.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.9 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.5 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 1.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.9 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.1 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 2.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.7 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 4.0 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 1.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.1 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.6 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 1.5 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 1.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.7 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 2.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.9 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.4 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.5 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.0 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.5 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.6 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 2.3 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 2.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.8 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 0.9 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 3.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 1.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.0 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 4.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 6.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 17.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.6 | 2.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 2.0 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.4 | 4.6 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 1.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.3 | 1.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 1.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 6.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 2.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 0.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.3 | 1.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.9 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.2 | 2.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 14.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 6.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 1.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 2.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 2.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 1.8 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 0.3 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.1 | 3.1 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 2.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.4 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 0.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.2 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 5.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 3.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 6.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.2 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.6 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 1.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 3.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.6 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 41.2 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 5.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 4.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.1 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.7 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 1.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 1.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 2.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |