Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
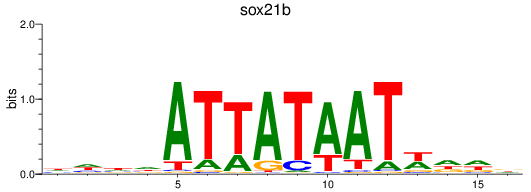
Results for sox21b
Z-value: 0.55
Transcription factors associated with sox21b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox21b
|
ENSDARG00000008540 | SRY-box transcription factor 21b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox21b | dr11_v1_chr9_+_53276356_53276356 | 0.13 | 7.3e-01 | Click! |
Activity profile of sox21b motif
Sorted Z-values of sox21b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_33154677 | 0.46 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr14_+_49135264 | 0.37 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr8_-_1838315 | 0.34 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr5_-_15494164 | 0.32 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr22_-_22337382 | 0.26 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr4_+_75200467 | 0.26 |
ENSDART00000122593
|
CABZ01043955.1
|
|
| chr22_+_12798569 | 0.25 |
ENSDART00000005720
|
stat1a
|
signal transducer and activator of transcription 1a |
| chr21_-_42007213 | 0.23 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr5_-_1203455 | 0.23 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr9_-_6661657 | 0.23 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr16_+_37582872 | 0.22 |
ENSDART00000169331
|
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr5_-_13167097 | 0.21 |
ENSDART00000149700
ENSDART00000030213 |
mapk1
|
mitogen-activated protein kinase 1 |
| chr18_+_924949 | 0.20 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr23_-_7125494 | 0.20 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr24_-_24271629 | 0.19 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr15_-_3534705 | 0.18 |
ENSDART00000158150
|
cog6
|
component of oligomeric golgi complex 6 |
| chr21_-_42007482 | 0.18 |
ENSDART00000075740
|
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_-_43915423 | 0.18 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr24_-_23784701 | 0.18 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr14_+_30285613 | 0.17 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr12_-_20796430 | 0.17 |
ENSDART00000064339
|
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr9_+_17971935 | 0.17 |
ENSDART00000149736
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr10_+_43188678 | 0.16 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr17_-_277046 | 0.16 |
ENSDART00000182587
|
LO018437.1
|
|
| chr9_-_1469083 | 0.16 |
ENSDART00000144096
|
rbm45
|
RNA binding motif protein 45 |
| chr7_+_26466826 | 0.16 |
ENSDART00000058908
|
mpdu1b
|
mannose-P-dolichol utilization defect 1b |
| chr13_-_24448278 | 0.16 |
ENSDART00000057584
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr11_-_42230491 | 0.16 |
ENSDART00000164423
|
CABZ01030862.1
|
|
| chr23_-_42810664 | 0.16 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr5_-_33825465 | 0.16 |
ENSDART00000110645
|
dab2ipb
|
DAB2 interacting protein b |
| chr11_-_26362294 | 0.16 |
ENSDART00000184654
ENSDART00000115037 |
foxj3
|
forkhead box J3 |
| chr10_+_35275965 | 0.16 |
ENSDART00000077404
|
pora
|
P450 (cytochrome) oxidoreductase a |
| chr5_-_67895656 | 0.16 |
ENSDART00000158917
|
abhd10a
|
abhydrolase domain containing 10a |
| chr18_+_9493720 | 0.16 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr5_+_27137473 | 0.16 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr10_-_20453995 | 0.16 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr16_-_42390441 | 0.15 |
ENSDART00000148475
|
cspg5a
|
chondroitin sulfate proteoglycan 5a |
| chr22_+_7439186 | 0.15 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr24_+_8904741 | 0.15 |
ENSDART00000140924
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr20_-_34028967 | 0.15 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr23_-_27050083 | 0.14 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr10_-_244745 | 0.14 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr23_+_40951443 | 0.14 |
ENSDART00000115161
|
reps2
|
RALBP1 associated Eps domain containing 2 |
| chr19_+_32401278 | 0.14 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
| chr25_-_12203952 | 0.14 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr11_-_1509773 | 0.14 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr17_+_3993772 | 0.14 |
ENSDART00000170822
ENSDART00000168613 ENSDART00000186174 |
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr19_+_30450125 | 0.14 |
ENSDART00000073704
|
si:ch211-215a10.4
|
si:ch211-215a10.4 |
| chr12_-_20796658 | 0.14 |
ENSDART00000181253
|
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr4_+_72742212 | 0.14 |
ENSDART00000171021
|
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr19_-_38872650 | 0.14 |
ENSDART00000146641
|
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr6_-_48473395 | 0.13 |
ENSDART00000185096
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr6_-_9563534 | 0.13 |
ENSDART00000184765
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr8_-_13678415 | 0.13 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr10_+_29204581 | 0.13 |
ENSDART00000148503
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr5_-_22868650 | 0.13 |
ENSDART00000057541
|
apool
|
apolipoprotein O-like |
| chr17_+_19630272 | 0.13 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr3_+_39540014 | 0.13 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr14_+_28281744 | 0.12 |
ENSDART00000173292
|
mid2
|
midline 2 |
| chr17_-_36936856 | 0.12 |
ENSDART00000010274
ENSDART00000188887 |
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr14_-_49896795 | 0.12 |
ENSDART00000168142
|
ube2d2l
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast), like |
| chr25_+_19913611 | 0.12 |
ENSDART00000155029
|
gramd4b
|
GRAM domain containing 4b |
| chr4_+_14899720 | 0.12 |
ENSDART00000187456
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr3_-_34296361 | 0.12 |
ENSDART00000181485
ENSDART00000113726 |
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr5_+_13521081 | 0.12 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr6_-_35310224 | 0.12 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr2_-_54387550 | 0.12 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr16_-_42872571 | 0.12 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr19_-_867071 | 0.11 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr8_+_32389838 | 0.11 |
ENSDART00000076350
ENSDART00000146901 |
mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2 like |
| chr12_-_9132682 | 0.11 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr20_-_34801181 | 0.11 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr17_-_8656155 | 0.11 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr6_-_57539141 | 0.11 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr25_+_4837915 | 0.11 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr5_+_54555567 | 0.10 |
ENSDART00000171159
|
anapc2
|
anaphase promoting complex subunit 2 |
| chr22_+_17399124 | 0.10 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr22_-_1245226 | 0.10 |
ENSDART00000161292
|
ampd2a
|
adenosine monophosphate deaminase 2a |
| chr12_-_37734973 | 0.10 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr20_-_7069612 | 0.10 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr13_-_19799824 | 0.10 |
ENSDART00000058032
|
fam204a
|
family with sequence similarity 204, member A |
| chr14_-_2327825 | 0.10 |
ENSDART00000180328
ENSDART00000191135 ENSDART00000114302 ENSDART00000189869 |
pcdh2ab8
pcdh2aa1
|
protocadherin 2 alpha b 8 protocadherin 2 alpha a 1 |
| chr22_+_883678 | 0.10 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr7_+_25135739 | 0.10 |
ENSDART00000167326
|
exoc6b
|
exocyst complex component 6B |
| chr12_+_4225199 | 0.10 |
ENSDART00000042277
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr5_-_33255759 | 0.10 |
ENSDART00000085531
|
prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr21_-_37951819 | 0.10 |
ENSDART00000139549
|
si:dkey-38k9.5
|
si:dkey-38k9.5 |
| chr6_+_48206535 | 0.09 |
ENSDART00000075172
|
cttnbp2nla
|
CTTNBP2 N-terminal like a |
| chr7_-_71434298 | 0.09 |
ENSDART00000180507
|
lgi2a
|
leucine-rich repeat LGI family, member 2a |
| chr21_+_38817785 | 0.09 |
ENSDART00000177616
ENSDART00000149085 |
hnf1bb
|
HNF1 homeobox Bb |
| chr12_+_46869271 | 0.09 |
ENSDART00000166560
|
hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr14_-_2217285 | 0.09 |
ENSDART00000157949
ENSDART00000166150 ENSDART00000054891 ENSDART00000183268 |
pcdh2ab2
pcdh2ab2
|
protocadherin 2 alpha b2 protocadherin 2 alpha b2 |
| chr7_+_69459759 | 0.09 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr19_-_42045372 | 0.09 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr14_+_35405518 | 0.09 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr3_-_32873641 | 0.09 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr7_+_30371893 | 0.08 |
ENSDART00000075513
|
aqp9b
|
aquaporin 9b |
| chr18_+_48423973 | 0.08 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr19_-_7450796 | 0.08 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr19_-_1363759 | 0.08 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr11_+_23993298 | 0.08 |
ENSDART00000186757
ENSDART00000172459 |
chia.1
|
chitinase, acidic.1 |
| chr23_+_25893020 | 0.08 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_+_29664336 | 0.08 |
ENSDART00000088290
|
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
| chr25_-_13549577 | 0.08 |
ENSDART00000166772
|
ano10b
|
anoctamin 10b |
| chr14_+_8456870 | 0.08 |
ENSDART00000007738
|
tmco6
|
transmembrane and coiled-coil domains 6 |
| chr17_+_15882533 | 0.08 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr25_+_37285737 | 0.08 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr4_+_9836465 | 0.08 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr6_+_41099787 | 0.08 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr18_+_13837746 | 0.07 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr19_-_22843480 | 0.07 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr10_-_34871737 | 0.07 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr1_+_41666611 | 0.07 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr2_+_11206317 | 0.07 |
ENSDART00000144982
|
lhx8a
|
LIM homeobox 8a |
| chr14_+_44545092 | 0.07 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr17_+_23146976 | 0.07 |
ENSDART00000114212
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_-_25464291 | 0.07 |
ENSDART00000112915
|
umad1
|
UBAP1-MVB12-associated (UMA) domain containing 1 |
| chr16_-_37372613 | 0.07 |
ENSDART00000124090
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr15_+_32711663 | 0.06 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr16_+_37470717 | 0.06 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr2_-_31791633 | 0.06 |
ENSDART00000180662
|
retreg1
|
reticulophagy regulator 1 |
| chr23_-_37291793 | 0.06 |
ENSDART00000083281
ENSDART00000187108 |
mul1b
|
mitochondrial E3 ubiquitin protein ligase 1b |
| chr2_+_44972720 | 0.06 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr23_+_42810055 | 0.06 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr1_+_34295925 | 0.06 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr25_+_36046287 | 0.06 |
ENSDART00000185324
|
rpgrip1l
|
RPGRIP1-like |
| chr17_+_16192486 | 0.06 |
ENSDART00000156832
|
kif13ba
|
kinesin family member 13Ba |
| chr21_+_39423974 | 0.06 |
ENSDART00000031470
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr1_-_46650022 | 0.06 |
ENSDART00000148759
ENSDART00000053222 |
rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr17_+_28533102 | 0.06 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr6_-_29086473 | 0.06 |
ENSDART00000124947
|
evi5b
|
ecotropic viral integration site 5b |
| chr20_-_16453602 | 0.06 |
ENSDART00000006443
|
gtf2a1
|
general transcription factor IIA, 1 |
| chr18_+_35842933 | 0.06 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr14_+_164556 | 0.06 |
ENSDART00000185606
|
WDR1
|
WD repeat domain 1 |
| chr12_-_15002563 | 0.06 |
ENSDART00000108852
ENSDART00000141909 |
pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr3_-_55537096 | 0.06 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr5_+_21144269 | 0.06 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr11_-_25733910 | 0.06 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr15_+_518092 | 0.05 |
ENSDART00000154594
|
pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr21_+_3854414 | 0.05 |
ENSDART00000122699
|
miga2
|
mitoguardin 2 |
| chr18_-_8313686 | 0.05 |
ENSDART00000182187
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr22_+_1170294 | 0.05 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr20_+_27712714 | 0.05 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr5_-_35161877 | 0.05 |
ENSDART00000139673
|
fcho2
|
FCH domain only 2 |
| chr17_+_44780166 | 0.05 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr22_+_16320076 | 0.05 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr11_+_21910343 | 0.05 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr11_-_33960318 | 0.05 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr19_+_2279051 | 0.05 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr25_-_27621268 | 0.05 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr2_-_24981356 | 0.05 |
ENSDART00000111212
|
nck1a
|
NCK adaptor protein 1a |
| chr11_-_42980535 | 0.05 |
ENSDART00000181160
ENSDART00000192064 |
CABZ01069998.1
|
|
| chr12_+_16967715 | 0.04 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr8_-_52473054 | 0.04 |
ENSDART00000193658
|
phyhip
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr21_-_29117670 | 0.04 |
ENSDART00000124327
|
havcr2
|
hepatitis A virus cellular receptor 2 |
| chr15_+_15314189 | 0.04 |
ENSDART00000156830
|
camkk1b
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha b |
| chr2_+_36109002 | 0.04 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr19_-_2876321 | 0.04 |
ENSDART00000159253
|
exosc7
|
exosome component 7 |
| chr8_+_25913787 | 0.04 |
ENSDART00000190257
ENSDART00000062515 |
sema3h
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3H |
| chr16_+_1100559 | 0.04 |
ENSDART00000092657
|
adamts16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr10_+_19595009 | 0.04 |
ENSDART00000112276
|
zgc:173837
|
zgc:173837 |
| chr8_-_31403938 | 0.04 |
ENSDART00000159168
|
nim1k
|
NIM1 serine/threonine protein kinase |
| chr12_-_19102736 | 0.04 |
ENSDART00000180364
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr11_-_34522249 | 0.04 |
ENSDART00000158616
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr24_-_32173754 | 0.04 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr10_-_15963903 | 0.04 |
ENSDART00000142357
|
si:dkey-3h23.3
|
si:dkey-3h23.3 |
| chr13_-_48764180 | 0.04 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr1_+_53714734 | 0.04 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr22_+_19552987 | 0.04 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr9_+_33222911 | 0.04 |
ENSDART00000135387
|
si:ch211-125e6.14
|
si:ch211-125e6.14 |
| chr19_-_28360033 | 0.04 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr1_+_32054159 | 0.04 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr22_-_38480186 | 0.04 |
ENSDART00000171704
|
soul4
|
heme-binding protein soul4 |
| chr2_+_56657804 | 0.04 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr16_-_44399335 | 0.04 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr18_+_29950233 | 0.04 |
ENSDART00000146431
|
atmin
|
ATM interactor |
| chr25_+_29662411 | 0.04 |
ENSDART00000077445
|
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr8_-_28304534 | 0.04 |
ENSDART00000078545
|
lmod1a
|
leiomodin 1a (smooth muscle) |
| chr2_+_39618951 | 0.04 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr24_+_21720304 | 0.03 |
ENSDART00000147250
ENSDART00000048273 |
pan3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr21_+_21263988 | 0.03 |
ENSDART00000089651
ENSDART00000108978 |
ccdc61
|
coiled-coil domain containing 61 |
| chr9_-_3519717 | 0.03 |
ENSDART00000145043
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr25_-_21782435 | 0.03 |
ENSDART00000089616
|
bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr11_-_10659195 | 0.03 |
ENSDART00000115255
|
mcf2l2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr13_+_24671481 | 0.03 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr13_-_43637371 | 0.03 |
ENSDART00000127930
ENSDART00000084474 |
fam160b1
|
family with sequence similarity 160, member B1 |
| chr6_-_43283122 | 0.03 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr4_+_31450683 | 0.03 |
ENSDART00000169303
|
znf996
|
zinc finger protein 996 |
| chr5_+_66353750 | 0.03 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr6_+_24398907 | 0.03 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr5_-_28767573 | 0.03 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr16_-_40841363 | 0.03 |
ENSDART00000163380
|
si:dkey-22o22.2
|
si:dkey-22o22.2 |
| chr20_-_34670236 | 0.03 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr10_+_42169982 | 0.03 |
ENSDART00000190905
|
CU467905.1
|
|
| chr22_+_33131891 | 0.03 |
ENSDART00000126885
|
dag1
|
dystroglycan 1 |
| chr15_-_23443937 | 0.03 |
ENSDART00000148311
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr6_-_1591002 | 0.03 |
ENSDART00000087039
|
zgc:123305
|
zgc:123305 |
| chr3_-_38745598 | 0.03 |
ENSDART00000127483
|
si:ch211-198m17.1
|
si:ch211-198m17.1 |
| chr11_-_44979281 | 0.03 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr2_-_19128385 | 0.03 |
ENSDART00000169793
|
si:dkey-225f23.3
|
si:dkey-225f23.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox21b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.2 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.0 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.2 | GO:0006003 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.4 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |