Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
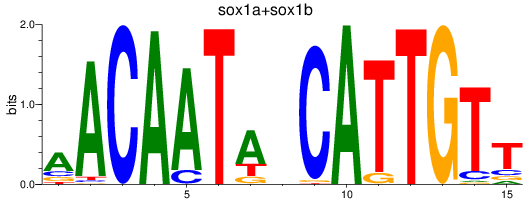
Results for sox1a+sox1b
Z-value: 1.07
Transcription factors associated with sox1a+sox1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox1b
|
ENSDARG00000008131 | SRY-box transcription factor 1b |
|
sox1a
|
ENSDARG00000069866 | SRY-box transcription factor 1a |
|
sox1a
|
ENSDARG00000110682 | SRY-box transcription factor 1a |
|
sox1a
|
ENSDARG00000114571 | SRY-box transcription factor 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox1b | dr11_v1_chr1_+_46194333_46194333 | 0.96 | 4.9e-05 | Click! |
| sox1a | dr11_v1_chr9_+_21722733_21722733 | 0.92 | 5.3e-04 | Click! |
Activity profile of sox1a+sox1b motif
Sorted Z-values of sox1a+sox1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_9421510 | 3.83 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr19_-_41472228 | 3.08 |
ENSDART00000113388
|
dlx5a
|
distal-less homeobox 5a |
| chr23_+_31815423 | 3.06 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr1_-_31856622 | 2.50 |
ENSDART00000065125
|
nt5c2b
|
5'-nucleotidase, cytosolic IIb |
| chr14_-_9420911 | 2.04 |
ENSDART00000081158
|
sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr18_-_23874929 | 1.67 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_51479128 | 1.61 |
ENSDART00000028018
|
meis1a
|
Meis homeobox 1 a |
| chr20_-_40370736 | 1.41 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr12_+_26512569 | 1.33 |
ENSDART00000066264
|
chad
|
chondroadherin |
| chr18_-_23875219 | 1.20 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_7715810 | 0.92 |
ENSDART00000130781
|
eif4a2
|
eukaryotic translation initiation factor 4A, isoform 2 |
| chr12_+_30168342 | 0.89 |
ENSDART00000142756
|
ablim1b
|
actin binding LIM protein 1b |
| chr7_+_61480296 | 0.84 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr14_-_17072736 | 0.84 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr16_-_50181206 | 0.81 |
ENSDART00000148478
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr19_+_11978209 | 0.81 |
ENSDART00000111568
|
polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr23_+_17800717 | 0.81 |
ENSDART00000122654
ENSDART00000044986 |
rnd1a
|
Rho family GTPase 1a |
| chr14_+_45883687 | 0.76 |
ENSDART00000114790
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr18_-_81280 | 0.74 |
ENSDART00000149810
ENSDART00000149321 |
hdc
|
histidine decarboxylase |
| chr14_+_36886950 | 0.73 |
ENSDART00000183719
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr2_-_53481912 | 0.70 |
ENSDART00000189610
|
hsd11b1lb
|
hydroxysteroid (11-beta) dehydrogenase 1-like b |
| chr17_-_37054959 | 0.64 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr16_-_8927425 | 0.62 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr19_+_18758014 | 0.62 |
ENSDART00000158935
|
clic1
|
chloride intracellular channel 1 |
| chr5_-_47727819 | 0.61 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr7_-_47148440 | 0.58 |
ENSDART00000185435
ENSDART00000189719 |
BX005073.3
|
|
| chr9_+_2333927 | 0.55 |
ENSDART00000123340
|
atp5mc3a
|
ATP synthase membrane subunit c locus 3a |
| chr22_+_10010292 | 0.48 |
ENSDART00000180096
|
BX324216.4
|
|
| chr24_+_35881984 | 0.47 |
ENSDART00000066587
|
klhl14
|
kelch-like family member 14 |
| chr11_+_37612672 | 0.45 |
ENSDART00000157993
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr7_+_57677120 | 0.44 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr24_-_9291857 | 0.41 |
ENSDART00000149875
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr12_-_4598237 | 0.41 |
ENSDART00000152489
|
irf3
|
interferon regulatory factor 3 |
| chrM_+_6425 | 0.40 |
ENSDART00000093606
|
mt-co1
|
cytochrome c oxidase I, mitochondrial |
| chr20_-_23852174 | 0.39 |
ENSDART00000122414
|
si:dkey-15j16.6
|
si:dkey-15j16.6 |
| chr23_-_44233408 | 0.38 |
ENSDART00000149318
ENSDART00000085528 |
zgc:158659
|
zgc:158659 |
| chr1_+_25732715 | 0.37 |
ENSDART00000140172
|
npy2r
|
neuropeptide Y receptor Y2 |
| chr10_-_34889053 | 0.36 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr14_-_3229384 | 0.34 |
ENSDART00000167247
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr15_-_951118 | 0.34 |
ENSDART00000171427
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr8_+_28065803 | 0.33 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr11_+_37612214 | 0.31 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr18_-_6766354 | 0.31 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr15_-_35410860 | 0.30 |
ENSDART00000191267
|
mecom
|
MDS1 and EVI1 complex locus |
| chr23_+_6926183 | 0.28 |
ENSDART00000149882
|
si:ch211-117c9.1
|
si:ch211-117c9.1 |
| chr15_-_2493771 | 0.25 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr7_+_1383027 | 0.24 |
ENSDART00000143762
|
khnyn
|
KH and NYN domain containing |
| chr9_-_6624663 | 0.24 |
ENSDART00000092537
|
GPR45
|
si:dkeyp-118h3.5 |
| chr18_-_4958883 | 0.23 |
ENSDART00000143614
|
zbbx
|
zinc finger, B-box domain containing |
| chr5_+_51833132 | 0.20 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr2_+_1881334 | 0.18 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr10_+_25726694 | 0.17 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr18_-_41754020 | 0.15 |
ENSDART00000077898
|
grk7b
|
G protein-coupled receptor kinase 7b |
| chr11_+_33284837 | 0.14 |
ENSDART00000166239
ENSDART00000111412 |
nkpd1
|
NTPase, KAP family P-loop domain containing 1 |
| chr5_+_51833305 | 0.12 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr19_+_9533008 | 0.12 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr19_+_23301050 | 0.10 |
ENSDART00000193713
|
si:dkey-79i2.4
|
si:dkey-79i2.4 |
| chr22_-_8931093 | 0.09 |
ENSDART00000181813
|
FO393424.3
|
|
| chr11_-_45434959 | 0.09 |
ENSDART00000173106
ENSDART00000172767 ENSDART00000172933 ENSDART00000172986 |
rfc4
|
replication factor C (activator 1) 4 |
| chr19_-_11977783 | 0.08 |
ENSDART00000183337
|
si:ch1073-296d18.1
|
si:ch1073-296d18.1 |
| chr25_-_19661198 | 0.07 |
ENSDART00000149641
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr11_+_16216909 | 0.06 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr2_-_54039293 | 0.06 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr7_-_71331499 | 0.06 |
ENSDART00000081245
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr7_-_71331690 | 0.05 |
ENSDART00000149682
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr8_-_436137 | 0.02 |
ENSDART00000187021
|
CABZ01063543.1
|
|
| chr7_+_50796422 | 0.01 |
ENSDART00000123868
|
dthd1
|
death domain containing 1 |
| chr22_+_9994833 | 0.01 |
ENSDART00000172696
ENSDART00000192101 |
BX324216.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of sox1a+sox1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.5 | 3.1 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.4 | 3.1 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 0.7 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 0.8 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.3 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.3 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.1 | 0.4 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.3 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.7 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.6 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.6 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 7.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 3.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.3 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 3.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |