Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
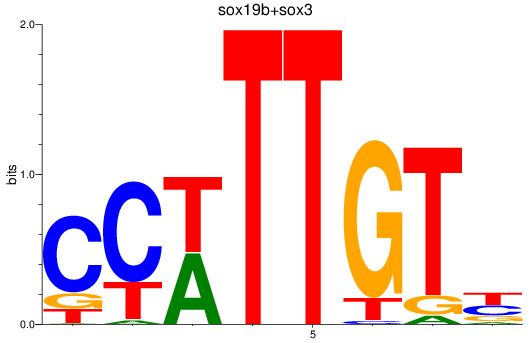
Results for sox19b+sox3
Z-value: 3.41
Transcription factors associated with sox19b+sox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox19b
|
ENSDARG00000040266 | SRY-box transcription factor 19b |
|
sox3
|
ENSDARG00000053569 | SRY-box transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox19b | dr11_v1_chr7_-_26497947_26497947 | -0.98 | 5.4e-06 | Click! |
| sox3 | dr11_v1_chr14_-_32744464_32744464 | 0.67 | 4.8e-02 | Click! |
Activity profile of sox19b+sox3 motif
Sorted Z-values of sox19b+sox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_44042033 | 11.09 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr10_+_33171501 | 9.92 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr24_-_40668208 | 6.35 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr9_-_22182396 | 6.02 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr6_+_52804267 | 5.87 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr14_-_36378494 | 5.80 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr1_-_49498116 | 5.39 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr9_-_22232902 | 5.35 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr9_-_22318511 | 5.33 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr13_+_22675802 | 5.22 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr9_-_22339582 | 5.15 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr9_-_22129788 | 5.11 |
ENSDART00000124272
ENSDART00000175417 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr7_+_39444843 | 5.04 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr14_+_46313396 | 5.04 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr1_+_44906517 | 4.99 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr12_+_5081759 | 4.97 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr10_+_18952271 | 4.97 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr16_+_34523515 | 4.94 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr14_+_46313135 | 4.86 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr21_+_27382893 | 4.85 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr9_-_22281854 | 4.72 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr9_-_22272181 | 4.70 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr20_+_31269778 | 4.60 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr9_-_22240052 | 4.54 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr5_+_35458190 | 4.52 |
ENSDART00000051313
|
fbp1b
|
fructose-1,6-bisphosphatase 1b |
| chr8_-_16697912 | 4.50 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr18_+_31016379 | 4.36 |
ENSDART00000172461
ENSDART00000163471 |
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr25_+_20089986 | 4.35 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr5_-_30615901 | 4.34 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr23_+_20110086 | 4.31 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr23_-_21471022 | 4.27 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr9_-_22188117 | 4.24 |
ENSDART00000132890
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr5_-_46505691 | 4.22 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr17_+_15433518 | 4.06 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr2_+_56463167 | 4.01 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr17_-_44247707 | 4.01 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr14_+_11457500 | 4.00 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr12_+_27129659 | 3.95 |
ENSDART00000076161
|
hoxb5b
|
homeobox B5b |
| chr6_+_12968101 | 3.87 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr19_-_17774875 | 3.87 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr17_+_15433671 | 3.84 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr19_+_12915498 | 3.80 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr10_+_9550419 | 3.74 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr11_-_41966854 | 3.72 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr14_-_1990290 | 3.67 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr7_-_7398350 | 3.64 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr25_+_35502552 | 3.64 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr19_+_24488403 | 3.64 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr5_-_36949476 | 3.63 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr9_-_11560427 | 3.62 |
ENSDART00000127942
ENSDART00000061442 |
cryba2b
|
crystallin, beta A2b |
| chr9_+_41103127 | 3.61 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr15_-_21877726 | 3.56 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr6_-_43092175 | 3.55 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr11_+_30295582 | 3.53 |
ENSDART00000122424
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr21_-_26495700 | 3.53 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr6_-_13783604 | 3.49 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr20_-_48485354 | 3.45 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr6_+_29791164 | 3.43 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr14_+_38786298 | 3.43 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr23_-_30787932 | 3.42 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr23_+_21459263 | 3.41 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr13_+_28854438 | 3.39 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr2_-_32768951 | 3.39 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr25_+_31323978 | 3.38 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr15_+_36115955 | 3.31 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr14_+_34486629 | 3.28 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr9_-_7684002 | 3.28 |
ENSDART00000016360
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr17_+_45305645 | 3.27 |
ENSDART00000172488
|
capn3a
|
calpain 3a, (p94) |
| chr9_-_7683799 | 3.25 |
ENSDART00000102713
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr20_-_43775495 | 3.24 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr9_+_54179306 | 3.24 |
ENSDART00000189829
|
tmsb4x
|
thymosin, beta 4 x |
| chr15_-_26549693 | 3.20 |
ENSDART00000186432
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr13_-_16257848 | 3.19 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr24_-_33756003 | 3.18 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr9_-_42696408 | 3.15 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr19_+_5315987 | 3.15 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr24_+_25069609 | 3.14 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr21_-_35853245 | 3.14 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr11_+_41981959 | 3.14 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr5_-_41531629 | 3.14 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr13_-_31441042 | 3.11 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr8_+_39619087 | 3.11 |
ENSDART00000134822
|
msi1
|
musashi RNA-binding protein 1 |
| chr10_+_21807497 | 3.05 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr22_-_10121880 | 3.04 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr16_-_24044664 | 3.03 |
ENSDART00000136982
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr18_-_15467446 | 3.03 |
ENSDART00000187847
|
endouc
|
endonuclease, polyU-specific C |
| chr9_+_32978302 | 3.01 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr7_-_69636502 | 3.00 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr23_-_10175898 | 2.97 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr3_-_30158395 | 2.93 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr7_-_24364536 | 2.93 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr23_+_8797143 | 2.92 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr21_+_25688388 | 2.90 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr14_-_41285392 | 2.87 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr4_-_16354292 | 2.86 |
ENSDART00000139919
|
lum
|
lumican |
| chr3_-_22212764 | 2.86 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr4_-_12862087 | 2.86 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr18_+_40525408 | 2.84 |
ENSDART00000132833
|
vwa7
|
von Willebrand factor A domain containing 7 |
| chr18_-_6633984 | 2.82 |
ENSDART00000185241
|
tnni1c
|
troponin I, skeletal, slow c |
| chr20_+_34770197 | 2.81 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr23_-_30045661 | 2.79 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr13_+_25428677 | 2.72 |
ENSDART00000186284
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr2_-_42960353 | 2.72 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr8_-_16712111 | 2.68 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr14_+_13454840 | 2.67 |
ENSDART00000161854
|
pls3
|
plastin 3 (T isoform) |
| chr8_+_23093155 | 2.66 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr25_+_34576067 | 2.65 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr16_+_23960933 | 2.65 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr16_+_23960744 | 2.65 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr5_-_65691512 | 2.64 |
ENSDART00000189255
|
notch1b
|
notch 1b |
| chr10_+_21780250 | 2.63 |
ENSDART00000183782
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr24_+_20559009 | 2.62 |
ENSDART00000131677
|
hhatlb
|
hedgehog acyltransferase like, b |
| chr7_-_28681724 | 2.61 |
ENSDART00000162400
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr25_-_204019 | 2.59 |
ENSDART00000188440
ENSDART00000191735 |
FP236318.2
|
|
| chr1_-_44434707 | 2.57 |
ENSDART00000110148
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr8_-_44904723 | 2.56 |
ENSDART00000040804
|
praf2
|
PRA1 domain family, member 2 |
| chr1_+_17376922 | 2.55 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr5_-_55560937 | 2.52 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr5_-_31875645 | 2.52 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr11_+_10548171 | 2.51 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr8_-_46509194 | 2.49 |
ENSDART00000038924
|
sult1st1
|
sulfotransferase family 1, cytosolic sulfotransferase 1 |
| chr14_-_2202652 | 2.47 |
ENSDART00000171316
|
si:dkey-262j3.7
|
si:dkey-262j3.7 |
| chr11_-_6048490 | 2.47 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr10_+_21783213 | 2.47 |
ENSDART00000168899
|
pcdh1g33
|
protocadherin 1 gamma 33 |
| chr10_-_28761454 | 2.47 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr19_+_10937932 | 2.46 |
ENSDART00000168968
|
si:ch73-347e22.8
|
si:ch73-347e22.8 |
| chr8_-_13541514 | 2.46 |
ENSDART00000063834
|
zgc:86586
|
zgc:86586 |
| chr15_-_3736149 | 2.45 |
ENSDART00000182986
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr9_+_30108641 | 2.43 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr11_+_8129536 | 2.43 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr14_-_2285955 | 2.43 |
ENSDART00000183928
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr1_+_46194333 | 2.40 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr11_+_14280598 | 2.39 |
ENSDART00000163033
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr11_-_42554290 | 2.37 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr6_+_43426599 | 2.36 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr14_-_26704829 | 2.36 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr7_+_20019125 | 2.35 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr9_-_22831836 | 2.35 |
ENSDART00000142585
|
neb
|
nebulin |
| chr14_-_8080416 | 2.34 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr18_-_6634424 | 2.33 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr20_+_26881600 | 2.33 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr23_-_42232124 | 2.33 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr23_+_45025909 | 2.33 |
ENSDART00000188105
|
hmgb2b
|
high mobility group box 2b |
| chr21_-_2310064 | 2.32 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr23_+_42813415 | 2.29 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr18_+_21122818 | 2.28 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr4_+_4232562 | 2.27 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr20_+_40237441 | 2.27 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr9_-_6380653 | 2.25 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr25_+_14017609 | 2.25 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr5_-_51756210 | 2.24 |
ENSDART00000163464
|
lhfpl2b
|
LHFPL tetraspan subfamily member 2b |
| chr9_-_1939232 | 2.23 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr12_-_7607114 | 2.22 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr19_+_19762183 | 2.22 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr23_-_24263474 | 2.22 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr19_-_41052408 | 2.21 |
ENSDART00000151120
|
sgce
|
sarcoglycan, epsilon |
| chr7_+_20524064 | 2.21 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr6_+_25261297 | 2.18 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr23_+_2669 | 2.17 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr4_+_5741733 | 2.15 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr20_-_20312789 | 2.15 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr8_-_16697615 | 2.15 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr5_+_37840914 | 2.12 |
ENSDART00000097738
|
panx1b
|
pannexin 1b |
| chr25_-_3830272 | 2.12 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr23_+_36144487 | 2.06 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr13_-_13294847 | 2.06 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr2_+_38161318 | 2.04 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr23_-_19715799 | 2.02 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr3_+_24134418 | 2.00 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr4_-_16345227 | 1.98 |
ENSDART00000079521
|
kera
|
keratocan |
| chr2_-_20599315 | 1.98 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr8_+_25351863 | 1.98 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr4_+_6643421 | 1.96 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr23_+_25428513 | 1.96 |
ENSDART00000144554
|
fmnl3
|
formin-like 3 |
| chr14_+_24277556 | 1.95 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr4_+_72742212 | 1.93 |
ENSDART00000171021
|
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr4_-_27301356 | 1.92 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr7_+_41295974 | 1.91 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr1_-_14233815 | 1.91 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr19_+_47394270 | 1.90 |
ENSDART00000171281
|
psmb2
|
proteasome subunit beta 2 |
| chr12_+_45677293 | 1.90 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr11_+_15613218 | 1.89 |
ENSDART00000066033
|
gdf11
|
growth differentiation factor 11 |
| chr5_+_15203421 | 1.89 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr21_-_20342096 | 1.88 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr6_-_3514529 | 1.88 |
ENSDART00000151020
ENSDART00000082151 |
uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr25_-_22187397 | 1.87 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr12_-_31009315 | 1.87 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7 like 2 |
| chr1_-_25936677 | 1.87 |
ENSDART00000146488
ENSDART00000136321 |
myoz2b
|
myozenin 2b |
| chr16_+_40576679 | 1.86 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr8_-_31938364 | 1.86 |
ENSDART00000137647
|
RNF180
|
si:dkey-250l23.4 |
| chr18_+_44649804 | 1.85 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr23_-_31506854 | 1.85 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr13_+_28821841 | 1.83 |
ENSDART00000179900
|
CU639469.1
|
|
| chr23_+_42819221 | 1.82 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr5_+_15667874 | 1.81 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr1_+_12767318 | 1.81 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr23_-_24148646 | 1.81 |
ENSDART00000113598
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr7_-_8408014 | 1.81 |
ENSDART00000112492
|
zgc:194686
|
zgc:194686 |
| chr1_-_26027327 | 1.81 |
ENSDART00000171292
ENSDART00000170878 |
si:ch211-145b13.6
|
si:ch211-145b13.6 |
| chr2_-_39017838 | 1.81 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox19b+sox3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.3 | GO:0016109 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 1.8 | 5.3 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 1.5 | 4.5 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 1.5 | 4.4 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.9 | 4.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.9 | 3.6 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.9 | 2.6 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.8 | 2.5 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.8 | 2.4 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.8 | 3.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.8 | 2.3 | GO:0035142 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.7 | 2.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.7 | 5.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 2.9 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.7 | 3.6 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.7 | 3.5 | GO:0043282 | pharyngeal muscle development(GO:0043282) |
| 0.7 | 4.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.7 | 2.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.7 | 6.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.6 | 1.9 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.6 | 3.7 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.6 | 1.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.6 | 79.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.6 | 6.7 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.5 | 1.6 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.5 | 3.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.5 | 3.1 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.5 | 3.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.5 | 2.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.5 | 1.9 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.5 | 1.4 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.5 | 1.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 3.5 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.4 | 2.2 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.4 | 2.6 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.4 | 2.5 | GO:0042745 | circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) |
| 0.4 | 2.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.4 | 2.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.4 | 8.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.4 | 2.2 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.4 | 2.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.4 | 1.8 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.4 | 6.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.4 | 3.9 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.4 | 5.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 3.8 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.3 | 1.0 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.3 | 1.4 | GO:0055016 | hypochord development(GO:0055016) |
| 0.3 | 1.7 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.3 | 1.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 2.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 13.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 1.5 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.3 | 2.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.3 | 1.2 | GO:0097037 | heme export(GO:0097037) |
| 0.3 | 5.0 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.3 | 1.2 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.3 | 3.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 1.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 1.0 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.3 | 1.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 2.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 1.2 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 2.7 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 1.4 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 1.3 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 1.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 3.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 1.4 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.2 | 2.7 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.2 | 1.9 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 2.7 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 1.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 0.5 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.2 | 1.8 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.2 | 0.4 | GO:0061213 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 0.9 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.3 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 | 2.5 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.2 | 1.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 1.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.2 | 1.9 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 2.5 | GO:0006584 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 1.0 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 2.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 1.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.7 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.4 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 33.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 1.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 2.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.6 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 0.6 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 1.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 2.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 2.7 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.6 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 3.0 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 0.9 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.7 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 2.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 1.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.0 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 4.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.8 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 3.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 3.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 2.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.3 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.1 | 1.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 3.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 2.9 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 3.8 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 1.4 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.1 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 2.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 3.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 2.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.5 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 1.3 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.1 | 1.8 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.4 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 2.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.3 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 1.2 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 1.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 2.7 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 5.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.9 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 3.1 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 2.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 1.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.1 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 2.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 4.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 4.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.5 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 1.6 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 1.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 1.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 1.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 7.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.3 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 2.3 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.6 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.5 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.0 | 1.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.0 | 1.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 1.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.3 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 1.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.0 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 2.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) multicellular organismal signaling(GO:0035637) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.7 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.8 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.6 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.3 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 1.2 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.5 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.2 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 2.3 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.4 | GO:0007596 | blood coagulation(GO:0007596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.3 | 3.9 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.9 | 6.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.9 | 4.6 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.8 | 3.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 3.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.6 | 7.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 1.9 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.5 | 1.9 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.5 | 1.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.5 | 19.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 3.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 6.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.1 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 0.6 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.2 | 3.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 2.9 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.2 | 1.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 1.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 2.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 3.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.9 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 4.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.6 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 1.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 3.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 7.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 6.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 5.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 5.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 12.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 6.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 3.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 3.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.3 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 1.8 | 5.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.5 | 4.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.3 | 3.9 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.2 | 3.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.0 | 77.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.8 | 2.5 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.8 | 3.9 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.8 | 4.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.7 | 3.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.6 | 3.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.6 | 6.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.6 | 10.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.5 | 2.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.5 | 7.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.5 | 1.5 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.5 | 3.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.5 | 2.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 1.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 1.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.4 | 3.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.4 | 1.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 1.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.4 | 2.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 1.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.3 | 1.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 2.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 2.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 1.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 5.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 1.6 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 1.9 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.3 | 1.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 2.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 1.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 1.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 1.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 0.6 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.2 | 1.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 1.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 1.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.9 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 5.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 3.1 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 1.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 3.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 2.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 6.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 3.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 0.5 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 2.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.1 | 1.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 3.1 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 3.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.6 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 2.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 2.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.9 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.7 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.6 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 1.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.9 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.8 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 2.4 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 0.4 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 2.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 3.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 8.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 70.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.4 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 3.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.7 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.0 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 3.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 5.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 17.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 2.0 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.3 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 60.6 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.4 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 3.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 3.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 6.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 11.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.5 | 6.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 4.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 2.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 5.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.3 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 2.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 1.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 2.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 1.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 1.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 2.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 5.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 1.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME TRANSLATION | Genes involved in Translation |