Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
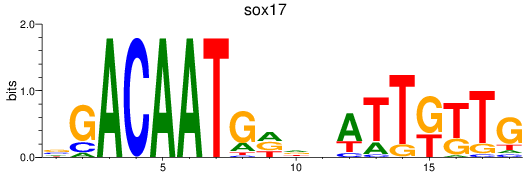
Results for sox17
Z-value: 0.82
Transcription factors associated with sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox17
|
ENSDARG00000101717 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox17 | dr11_v1_chr7_-_58776400_58776400 | -0.53 | 1.4e-01 | Click! |
Activity profile of sox17 motif
Sorted Z-values of sox17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_20560616 | 1.92 |
ENSDART00000136710
ENSDART00000151974 ENSDART00000121699 ENSDART00000040074 |
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr5_-_15264007 | 1.09 |
ENSDART00000180641
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr6_+_54498220 | 1.02 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr16_+_50741154 | 0.96 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr8_-_44611357 | 0.88 |
ENSDART00000063396
|
bag4
|
BCL2 associated athanogene 4 |
| chr9_-_16218097 | 0.83 |
ENSDART00000190503
|
myo1b
|
myosin IB |
| chr1_+_53321878 | 0.81 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr3_+_18437397 | 0.81 |
ENSDART00000136243
ENSDART00000184882 ENSDART00000135470 |
tbc1d16
|
TBC1 domain family, member 16 |
| chr13_-_35765028 | 0.78 |
ENSDART00000157391
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr22_+_25274712 | 0.75 |
ENSDART00000137341
|
BX950205.1
|
|
| chr17_+_16423721 | 0.73 |
ENSDART00000064233
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr24_-_10021341 | 0.72 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr24_+_31459715 | 0.72 |
ENSDART00000181102
ENSDART00000189950 ENSDART00000192321 ENSDART00000126380 |
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr8_+_13368150 | 0.72 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr23_+_32101361 | 0.72 |
ENSDART00000138849
|
zgc:56699
|
zgc:56699 |
| chr12_-_30359498 | 0.71 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr12_-_30358836 | 0.70 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr12_-_30359031 | 0.70 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr19_+_42886413 | 0.70 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr11_-_42134968 | 0.69 |
ENSDART00000187115
|
FP325130.1
|
|
| chr9_+_16854121 | 0.69 |
ENSDART00000110866
|
cln5
|
CLN5, intracellular trafficking protein |
| chr3_-_55525627 | 0.69 |
ENSDART00000189234
|
tex2
|
testis expressed 2 |
| chr5_-_20678300 | 0.68 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr3_+_35005730 | 0.68 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr17_-_12058171 | 0.68 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr17_-_2584423 | 0.68 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr19_+_42432625 | 0.67 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr11_-_27537593 | 0.66 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr15_-_18209672 | 0.66 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr16_-_41717063 | 0.65 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr18_+_17534627 | 0.64 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr2_-_26596794 | 0.64 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr6_+_30533504 | 0.64 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr15_-_28904371 | 0.64 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr14_-_24101897 | 0.61 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr2_-_17492486 | 0.61 |
ENSDART00000189464
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr10_+_15454745 | 0.60 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr10_-_3258073 | 0.60 |
ENSDART00000113162
|
pi4kaa
|
phosphatidylinositol 4-kinase, catalytic, alpha a |
| chr1_-_46650022 | 0.59 |
ENSDART00000148759
ENSDART00000053222 |
rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr9_-_46276626 | 0.59 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr2_-_17492080 | 0.58 |
ENSDART00000024302
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr4_-_3145359 | 0.58 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr20_-_45062514 | 0.57 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr24_+_10027902 | 0.57 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr17_+_13031497 | 0.57 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr23_+_42254960 | 0.56 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr23_+_16774589 | 0.56 |
ENSDART00000104791
|
zgc:153722
|
zgc:153722 |
| chr21_+_12036238 | 0.55 |
ENSDART00000102463
ENSDART00000155426 |
zgc:162344
|
zgc:162344 |
| chr5_+_37504309 | 0.55 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr16_-_31598771 | 0.55 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr21_-_39546737 | 0.54 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr22_+_14051245 | 0.54 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr20_-_45060241 | 0.54 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr24_+_26406770 | 0.53 |
ENSDART00000110011
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr25_+_3104959 | 0.53 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr5_-_8602937 | 0.53 |
ENSDART00000157765
|
rictora
|
RPTOR independent companion of MTOR, complex 2 a |
| chr5_-_30074332 | 0.52 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr13_+_31687973 | 0.52 |
ENSDART00000076479
|
slc38a6
|
solute carrier family 38, member 6 |
| chr12_-_9123352 | 0.52 |
ENSDART00000149655
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr20_-_18794789 | 0.51 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr10_-_45029041 | 0.51 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr7_-_30624435 | 0.50 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr11_-_33868881 | 0.50 |
ENSDART00000163295
ENSDART00000172633 ENSDART00000171439 |
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr3_-_12381271 | 0.49 |
ENSDART00000171068
ENSDART00000157475 |
coro7
|
coronin 7 |
| chr25_+_20694177 | 0.49 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr7_+_23495986 | 0.48 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr4_-_18416566 | 0.48 |
ENSDART00000033717
|
cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr23_+_19655301 | 0.48 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr17_+_24222190 | 0.48 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr7_+_34592526 | 0.48 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr18_-_21725805 | 0.47 |
ENSDART00000182185
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr23_-_24542156 | 0.46 |
ENSDART00000132265
|
atp13a2
|
ATPase 13A2 |
| chr19_-_25114701 | 0.46 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr16_-_33095161 | 0.46 |
ENSDART00000187648
|
dopey1
|
dopey family member 1 |
| chr4_+_69801372 | 0.45 |
ENSDART00000154125
|
si:dkey-238o14.7
|
si:dkey-238o14.7 |
| chr12_-_611367 | 0.45 |
ENSDART00000152286
|
wu:fj29h11
|
wu:fj29h11 |
| chr4_-_5370859 | 0.45 |
ENSDART00000189559
|
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr21_-_2248239 | 0.45 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr7_-_32020100 | 0.45 |
ENSDART00000185433
|
kif18a
|
kinesin family member 18A |
| chr17_-_23895026 | 0.45 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr23_+_30730121 | 0.45 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr8_-_22326073 | 0.45 |
ENSDART00000084965
|
cep104
|
centrosomal protein 104 |
| chr21_+_26748141 | 0.44 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr7_+_38349667 | 0.44 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr4_-_2975461 | 0.44 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr18_-_21725638 | 0.44 |
ENSDART00000089853
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr6_+_4299164 | 0.43 |
ENSDART00000159759
|
nbeal1
|
neurobeachin-like 1 |
| chr11_-_44979281 | 0.43 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr22_+_14051894 | 0.43 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr21_+_6328801 | 0.43 |
ENSDART00000163577
|
fnbp1b
|
formin binding protein 1b |
| chr14_+_26566936 | 0.43 |
ENSDART00000173056
|
arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr18_+_44630415 | 0.43 |
ENSDART00000098540
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr8_+_10339869 | 0.42 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr22_-_27113332 | 0.42 |
ENSDART00000146951
ENSDART00000178855 |
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr20_+_18945057 | 0.42 |
ENSDART00000035447
|
mtmr9
|
myotubularin related protein 9 |
| chr10_+_4235998 | 0.42 |
ENSDART00000169328
|
plppr1
|
phospholipid phosphatase related 1 |
| chr1_-_23110740 | 0.42 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr22_+_12361489 | 0.42 |
ENSDART00000182483
|
r3hdm1
|
R3H domain containing 1 |
| chr16_-_28658341 | 0.42 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr22_+_32228882 | 0.41 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr6_+_6828167 | 0.41 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr24_+_26997798 | 0.41 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr20_-_31238313 | 0.41 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr12_-_3077395 | 0.40 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr2_+_22409249 | 0.40 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr7_+_66884570 | 0.40 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr5_-_57289872 | 0.40 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr16_-_30826712 | 0.40 |
ENSDART00000122474
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr22_+_12361317 | 0.40 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr15_-_30815826 | 0.40 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr2_+_43204919 | 0.39 |
ENSDART00000160077
ENSDART00000018729 ENSDART00000129134 ENSDART00000056402 |
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr17_-_20218167 | 0.39 |
ENSDART00000154479
|
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr9_-_6618067 | 0.39 |
ENSDART00000061585
|
si:dkeyp-118h3.6
|
si:dkeyp-118h3.6 |
| chr6_-_54444929 | 0.39 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr21_+_21791343 | 0.39 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr6_-_39198912 | 0.38 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr23_+_34321237 | 0.38 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr2_+_36109002 | 0.38 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr6_+_29923593 | 0.37 |
ENSDART00000169687
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr6_-_37513133 | 0.37 |
ENSDART00000147826
|
jade3
|
jade family PHD finger 3 |
| chr3_-_40976463 | 0.37 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr23_+_24973773 | 0.37 |
ENSDART00000047020
|
casp9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr9_-_22057658 | 0.37 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr19_+_20495249 | 0.37 |
ENSDART00000090810
|
kcnh8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr11_+_25539698 | 0.37 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr1_-_18772241 | 0.37 |
ENSDART00000147645
ENSDART00000133194 ENSDART00000147525 ENSDART00000191493 |
tbc1d1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr24_-_16979728 | 0.37 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr15_+_20344670 | 0.36 |
ENSDART00000158986
|
plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr11_-_45385803 | 0.36 |
ENSDART00000173329
|
trappc10
|
trafficking protein particle complex 10 |
| chr4_-_3064101 | 0.36 |
ENSDART00000135701
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr24_+_5829265 | 0.36 |
ENSDART00000155242
|
si:ch211-157j23.5
|
si:ch211-157j23.5 |
| chr11_+_30636351 | 0.36 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr3_+_35611625 | 0.36 |
ENSDART00000190995
|
traf7
|
TNF receptor-associated factor 7 |
| chr21_-_32781612 | 0.36 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr1_+_46405294 | 0.35 |
ENSDART00000143228
|
tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr7_-_18601206 | 0.35 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr14_+_16093889 | 0.35 |
ENSDART00000159181
|
immt
|
inner membrane protein, mitochondrial (mitofilin) |
| chr5_-_67349916 | 0.35 |
ENSDART00000144092
|
mlxip
|
MLX interacting protein |
| chr16_-_54978981 | 0.35 |
ENSDART00000154023
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr19_-_20446756 | 0.35 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr6_-_39199070 | 0.34 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr7_+_21917857 | 0.34 |
ENSDART00000088041
|
ufsp1
|
UFM1-specific peptidase 1 (non-functional) |
| chr10_-_28027490 | 0.34 |
ENSDART00000185445
|
ints2
|
integrator complex subunit 2 |
| chr5_-_69314495 | 0.34 |
ENSDART00000182335
|
smtnb
|
smoothelin b |
| chr3_+_28860283 | 0.34 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr2_+_24867534 | 0.34 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr12_-_30032188 | 0.34 |
ENSDART00000042514
|
atrnl1b
|
attractin-like 1b |
| chr20_+_4221978 | 0.33 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr18_+_31117136 | 0.33 |
ENSDART00000138403
|
tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr17_-_20218357 | 0.33 |
ENSDART00000155990
ENSDART00000155632 ENSDART00000156540 ENSDART00000063523 |
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr12_+_19958845 | 0.33 |
ENSDART00000193248
|
ercc4
|
excision repair cross-complementation group 4 |
| chr7_-_28641522 | 0.33 |
ENSDART00000182071
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr24_-_25030173 | 0.33 |
ENSDART00000126395
ENSDART00000019350 |
nup58
|
nucleoporin 58 |
| chr18_+_31117323 | 0.32 |
ENSDART00000059893
|
tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr1_+_46404968 | 0.32 |
ENSDART00000042064
|
tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr4_-_28353538 | 0.32 |
ENSDART00000064219
|
trmu
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr4_-_12781182 | 0.31 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr9_+_38457806 | 0.31 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr21_+_33503835 | 0.31 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr13_-_533243 | 0.31 |
ENSDART00000038315
|
nvl
|
nuclear VCP-like |
| chr24_-_27409599 | 0.31 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr6_-_7842078 | 0.31 |
ENSDART00000065507
|
plppr2b
|
phospholipid phosphatase related 2b |
| chr16_-_11858009 | 0.31 |
ENSDART00000158347
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr24_-_12832499 | 0.31 |
ENSDART00000182027
ENSDART00000039312 |
ipo4
|
importin 4 |
| chr7_-_71384391 | 0.31 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr19_+_43905671 | 0.30 |
ENSDART00000168725
ENSDART00000133628 |
ankib1a
|
ankyrin repeat and IBR domain containing 1a |
| chr14_+_29609245 | 0.30 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr7_+_9041174 | 0.30 |
ENSDART00000163411
|
AL935128.1
|
|
| chr11_-_1948784 | 0.30 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr23_-_36670369 | 0.30 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| chr2_+_55199721 | 0.30 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr20_-_48898371 | 0.30 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr24_-_25673405 | 0.30 |
ENSDART00000186081
ENSDART00000110241 ENSDART00000142351 |
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr18_+_1837668 | 0.30 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr5_-_22868650 | 0.30 |
ENSDART00000057541
|
apool
|
apolipoprotein O-like |
| chr19_-_24136233 | 0.29 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr17_-_6599484 | 0.29 |
ENSDART00000156927
|
ANKRD66
|
si:ch211-189e2.2 |
| chr10_-_6588793 | 0.29 |
ENSDART00000163788
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr16_-_25400257 | 0.29 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr15_-_43768776 | 0.29 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr5_-_26323137 | 0.28 |
ENSDART00000133823
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr22_+_1352192 | 0.28 |
ENSDART00000169629
|
znf45l
|
zinc finger 45 like |
| chr23_+_26017227 | 0.28 |
ENSDART00000002939
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr6_-_7107868 | 0.28 |
ENSDART00000171123
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr14_-_46113321 | 0.28 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr2_+_51818039 | 0.28 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr13_-_31687925 | 0.27 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr23_-_33702900 | 0.27 |
ENSDART00000187413
|
pou6f1
|
POU class 6 homeobox 1 |
| chr1_-_56951369 | 0.27 |
ENSDART00000152294
|
si:ch211-1f22.2
|
si:ch211-1f22.2 |
| chr5_-_5243079 | 0.27 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr13_-_48431766 | 0.27 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr8_+_42717005 | 0.27 |
ENSDART00000181656
ENSDART00000180320 ENSDART00000181293 |
zgc:194007
|
zgc:194007 |
| chr5_-_33769211 | 0.27 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr5_+_36530836 | 0.27 |
ENSDART00000006899
|
igbp1
|
immunoglobulin (CD79A) binding protein 1 |
| chr16_-_47301376 | 0.26 |
ENSDART00000169697
|
mios
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr23_-_45319592 | 0.26 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr3_+_15773991 | 0.26 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr21_-_21530868 | 0.26 |
ENSDART00000174231
|
or133-9
|
odorant receptor, family H, subfamily 133, member 9 |
| chr7_+_5936601 | 0.26 |
ENSDART00000181693
|
si:dkey-23a13.11
|
si:dkey-23a13.11 |
| chr7_+_34297271 | 0.26 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.2 | 2.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.2 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.6 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.6 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.5 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.3 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.3 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.9 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.4 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.3 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 0.6 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 0.3 | GO:0019068 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 0.2 | GO:0071498 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.5 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.5 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.6 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.5 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.6 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.7 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.8 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.6 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.4 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.7 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.4 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.9 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 2.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.7 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.0 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.3 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.4 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) hypothalamus cell differentiation(GO:0021979) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 0.7 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 0.5 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.6 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 0.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.5 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 0.9 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.3 | 2.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.4 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.7 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 2.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.9 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |