Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
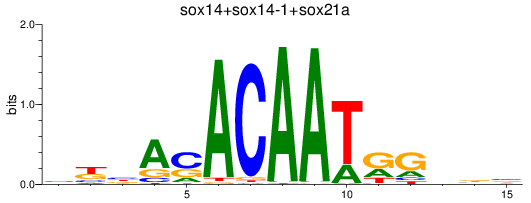
Results for sox14+sox14-1+sox21a
Z-value: 1.18
Transcription factors associated with sox14+sox14-1+sox21a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox21a
|
ENSDARG00000031664 | SRY-box transcription factor 21a |
|
sox14
|
ENSDARG00000070929 | SRY-box transcription factor 14 |
|
sox21a
|
ENSDARG00000111842 | SRY-box transcription factor 21a |
|
sox14-1
|
ENSDARG00000111969 | SRY-box transcription factor 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox14 | dr11_v1_chr6_-_26559921_26559921 | 0.68 | 4.4e-02 | Click! |
| FQ377628.2 | dr11_v1_chr2_-_22575851_22575851 | -0.36 | 3.4e-01 | Click! |
| sox21a | dr11_v1_chr6_+_7414215_7414215 | 0.18 | 6.4e-01 | Click! |
Activity profile of sox14+sox14-1+sox21a motif
Sorted Z-values of sox14+sox14-1+sox21a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_46313396 | 2.52 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr14_+_46313135 | 2.03 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr9_-_11560110 | 1.80 |
ENSDART00000176941
|
cryba2b
|
crystallin, beta A2b |
| chr23_-_6641223 | 1.78 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr10_+_44042033 | 1.76 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr21_+_26390549 | 1.52 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr5_-_41531629 | 1.24 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr5_-_30615901 | 1.22 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr9_-_11560427 | 1.19 |
ENSDART00000127942
ENSDART00000061442 |
cryba2b
|
crystallin, beta A2b |
| chr6_-_39764995 | 1.12 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_+_31891110 | 1.07 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr12_+_17865374 | 1.01 |
ENSDART00000169019
|
tmem130
|
transmembrane protein 130 |
| chr6_-_39765546 | 0.89 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr2_-_39017838 | 0.78 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr10_+_33171501 | 0.75 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr14_-_25599002 | 0.73 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr23_+_36130883 | 0.73 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr13_-_31470439 | 0.70 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr8_-_17067364 | 0.68 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr21_-_16113799 | 0.65 |
ENSDART00000187848
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr2_-_6112862 | 0.63 |
ENSDART00000164269
|
prdx1
|
peroxiredoxin 1 |
| chr12_+_22576404 | 0.62 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr1_+_26356360 | 0.61 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr9_-_23922011 | 0.61 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr7_+_25913225 | 0.60 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr21_-_16113477 | 0.59 |
ENSDART00000147588
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr10_+_18952271 | 0.58 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr12_-_4346085 | 0.57 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr4_-_27301356 | 0.57 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr23_+_42819221 | 0.55 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr9_-_23922778 | 0.51 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr3_-_25814097 | 0.47 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr15_-_21132480 | 0.46 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr14_+_11457500 | 0.45 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr17_-_37214196 | 0.45 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr14_-_2189889 | 0.44 |
ENSDART00000181557
ENSDART00000106707 |
pcdh2ab9
pcdh2ab11
|
protocadherin 2 alpha b 9 protocadherin 2 alpha b 11 |
| chr8_+_18830759 | 0.44 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr11_+_15613218 | 0.44 |
ENSDART00000066033
|
gdf11
|
growth differentiation factor 11 |
| chr18_-_38087875 | 0.43 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr1_-_35695614 | 0.43 |
ENSDART00000133813
ENSDART00000145291 ENSDART00000176451 |
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr21_+_43669943 | 0.43 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr2_-_56348727 | 0.42 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr16_-_51888952 | 0.41 |
ENSDART00000186407
ENSDART00000175435 |
rpl28
|
ribosomal protein L28 |
| chr18_+_46151505 | 0.41 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr6_+_43426599 | 0.41 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr17_-_36929332 | 0.39 |
ENSDART00000183454
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr4_+_25950372 | 0.39 |
ENSDART00000125767
|
metap2a
|
methionyl aminopeptidase 2a |
| chr16_+_50089417 | 0.39 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr1_+_26356205 | 0.39 |
ENSDART00000190064
ENSDART00000176380 |
tet2
|
tet methylcytosine dioxygenase 2 |
| chr8_-_13541514 | 0.38 |
ENSDART00000063834
|
zgc:86586
|
zgc:86586 |
| chr23_-_19715799 | 0.38 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr5_-_26834511 | 0.37 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr15_-_36533322 | 0.36 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr8_+_39619087 | 0.36 |
ENSDART00000134822
|
msi1
|
musashi RNA-binding protein 1 |
| chr2_+_6963296 | 0.36 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr21_-_26071142 | 0.36 |
ENSDART00000004740
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr24_-_40700596 | 0.35 |
ENSDART00000162635
|
smyhc2
|
slow myosin heavy chain 2 |
| chr14_+_9462726 | 0.35 |
ENSDART00000063559
|
rell2
|
RELT-like 2 |
| chr6_+_584632 | 0.35 |
ENSDART00000151150
|
zgc:92360
|
zgc:92360 |
| chr4_-_17805128 | 0.34 |
ENSDART00000128988
|
spi2
|
Spi-2 proto-oncogene |
| chr10_+_21722892 | 0.34 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr12_-_431249 | 0.33 |
ENSDART00000083827
|
hs3st3l
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3-like |
| chr23_+_42813415 | 0.33 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr1_+_59088205 | 0.32 |
ENSDART00000150649
ENSDART00000100197 |
zgc:173915
|
zgc:173915 |
| chr17_-_32863250 | 0.32 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr1_-_49225890 | 0.32 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr16_-_24703603 | 0.31 |
ENSDART00000131167
|
negaly6
|
neuromast-expressed gpi-anchored lymphocyte antigen 6 |
| chr18_+_21122818 | 0.31 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr19_+_47311020 | 0.31 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr11_+_14280598 | 0.31 |
ENSDART00000163033
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr8_-_49172418 | 0.29 |
ENSDART00000142437
ENSDART00000036157 |
fkbp1aa
|
FK506 binding protein 1Aa |
| chr17_-_46457622 | 0.28 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr13_+_30912385 | 0.27 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr5_+_36752943 | 0.27 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr23_-_27857051 | 0.27 |
ENSDART00000043462
ENSDART00000137861 |
acvrl1
|
activin A receptor like type 1 |
| chr9_-_51436377 | 0.27 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr4_-_240737 | 0.26 |
ENSDART00000166186
|
RERG
|
si:cabz01085950.1 |
| chr12_+_22680115 | 0.25 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr22_-_15693085 | 0.25 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr25_+_10793478 | 0.24 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr5_-_57528943 | 0.24 |
ENSDART00000130320
|
pisd
|
phosphatidylserine decarboxylase |
| chr10_+_32560667 | 0.23 |
ENSDART00000182278
|
map6a
|
microtubule-associated protein 6a |
| chr12_-_16084835 | 0.22 |
ENSDART00000090881
|
kcnj19b
|
potassium voltage-gated channel, subfamily J, member 19b |
| chr9_-_21838045 | 0.22 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr22_+_1028724 | 0.22 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr10_-_342564 | 0.21 |
ENSDART00000157633
|
vaspa
|
vasodilator stimulated phosphoprotein a |
| chr10_-_27223827 | 0.20 |
ENSDART00000185138
|
auts2a
|
autism susceptibility candidate 2a |
| chr3_-_28665291 | 0.20 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr7_-_1965927 | 0.20 |
ENSDART00000053656
|
si:cabz01040626.2
|
si:cabz01040626.2 |
| chr19_-_7420867 | 0.20 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr24_-_27400017 | 0.20 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr8_-_45760087 | 0.19 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr10_-_43568239 | 0.19 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr7_+_31145386 | 0.19 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr22_-_13857729 | 0.19 |
ENSDART00000177971
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr18_+_14477740 | 0.18 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr6_+_12968101 | 0.18 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr5_-_201600 | 0.18 |
ENSDART00000158495
|
CABZ01088906.1
|
|
| chr4_-_64123545 | 0.17 |
ENSDART00000170040
|
BX914205.2
|
|
| chr19_-_41404870 | 0.17 |
ENSDART00000167772
|
sem1
|
SEM1, 26S proteasome complex subunit |
| chr20_+_88168 | 0.17 |
ENSDART00000149283
|
zgc:112001
|
zgc:112001 |
| chr16_-_24703855 | 0.17 |
ENSDART00000182422
|
negaly6
|
neuromast-expressed gpi-anchored lymphocyte antigen 6 |
| chr5_-_26181863 | 0.16 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr12_-_17479078 | 0.16 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr25_+_26844028 | 0.16 |
ENSDART00000127274
ENSDART00000156179 |
sema7a
|
semaphorin 7A |
| chr19_-_3916075 | 0.16 |
ENSDART00000161799
|
si:ch73-281i18.7
|
si:ch73-281i18.7 |
| chr25_+_32474031 | 0.15 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr1_-_55118745 | 0.15 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr7_+_9922607 | 0.15 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr7_-_27696958 | 0.15 |
ENSDART00000173470
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr6_+_7466223 | 0.15 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr8_-_46457233 | 0.15 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr13_+_7292061 | 0.15 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr14_-_38873095 | 0.15 |
ENSDART00000047050
ENSDART00000173285 ENSDART00000147521 |
gsr
|
glutathione reductase |
| chr11_+_583142 | 0.14 |
ENSDART00000168157
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr19_-_5125663 | 0.14 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr7_+_20019125 | 0.13 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr24_-_21914276 | 0.13 |
ENSDART00000128687
|
c1qtnf9
|
C1q and TNF related 9 |
| chr13_+_30912938 | 0.12 |
ENSDART00000190003
|
drgx
|
dorsal root ganglia homeobox |
| chr11_-_30158191 | 0.12 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr19_-_18418763 | 0.12 |
ENSDART00000167271
|
zgc:112966
|
zgc:112966 |
| chr24_-_26485098 | 0.12 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr23_-_7826849 | 0.12 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr11_+_36665359 | 0.11 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr21_-_29100110 | 0.11 |
ENSDART00000142598
|
timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr1_+_54865552 | 0.11 |
ENSDART00000145381
|
si:ch211-196h16.5
|
si:ch211-196h16.5 |
| chr20_+_32152355 | 0.11 |
ENSDART00000152904
ENSDART00000139507 |
sesn1
|
sestrin 1 |
| chr17_+_14711765 | 0.11 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr10_+_36197968 | 0.11 |
ENSDART00000114102
|
COA4
|
si:ch211-215a9.5 |
| chr6_+_4229360 | 0.11 |
ENSDART00000191347
ENSDART00000130642 |
FO082877.1
|
|
| chr3_+_27713610 | 0.11 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr9_+_53707240 | 0.11 |
ENSDART00000171490
|
PCDH8
|
si:ch211-199f5.1 |
| chr1_+_56755536 | 0.10 |
ENSDART00000157727
|
CABZ01049361.1
|
|
| chr8_-_46486009 | 0.10 |
ENSDART00000140431
|
sult1st9
|
sulfotransferase family 1, cytosolic sulfotransferase 9 |
| chr2_-_51521079 | 0.10 |
ENSDART00000165504
|
LO018493.1
|
|
| chr19_+_46222918 | 0.10 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr21_+_24984970 | 0.10 |
ENSDART00000088546
|
jam3b
|
junctional adhesion molecule 3b |
| chr25_+_20081553 | 0.10 |
ENSDART00000174684
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr8_-_30204650 | 0.10 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr7_+_65876335 | 0.10 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr14_-_29906209 | 0.09 |
ENSDART00000192952
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr2_-_51512294 | 0.09 |
ENSDART00000185287
|
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr7_+_8324506 | 0.09 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr13_-_37636147 | 0.08 |
ENSDART00000144065
|
si:dkey-188i13.8
|
si:dkey-188i13.8 |
| chr5_-_66028371 | 0.08 |
ENSDART00000183012
|
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr21_+_32820175 | 0.08 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr25_-_29134654 | 0.08 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr22_-_10019768 | 0.08 |
ENSDART00000162077
|
BX324216.2
|
|
| chr11_+_583725 | 0.07 |
ENSDART00000189415
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr4_-_73739119 | 0.07 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr6_+_10338554 | 0.07 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr23_-_35491587 | 0.07 |
ENSDART00000153794
|
fam110c
|
family with sequence similarity 110, member C |
| chr3_-_50139860 | 0.07 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr8_+_52503340 | 0.07 |
ENSDART00000168838
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr13_-_13294847 | 0.07 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr12_+_19030391 | 0.07 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr19_+_46222428 | 0.07 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr12_+_30046320 | 0.07 |
ENSDART00000179904
ENSDART00000153394 |
ablim1b
|
actin binding LIM protein 1b |
| chr13_-_20519001 | 0.07 |
ENSDART00000168955
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr4_+_5392094 | 0.07 |
ENSDART00000135122
|
si:dkey-14d8.20
|
si:dkey-14d8.20 |
| chr11_-_41130239 | 0.07 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr9_-_24244383 | 0.06 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr14_-_21238046 | 0.06 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr7_+_35268880 | 0.06 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr3_-_19091024 | 0.06 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr14_-_3268155 | 0.06 |
ENSDART00000177244
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr11_-_16093018 | 0.05 |
ENSDART00000139309
ENSDART00000139819 |
SPATA1
|
si:dkey-205k8.5 |
| chr12_-_44010532 | 0.05 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr3_-_18675688 | 0.05 |
ENSDART00000048218
|
slc5a11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr6_+_30456788 | 0.05 |
ENSDART00000121492
|
FP236735.1
|
|
| chr1_-_18848955 | 0.05 |
ENSDART00000109294
ENSDART00000146410 |
zgc:195282
|
zgc:195282 |
| chr7_-_72426484 | 0.04 |
ENSDART00000190063
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr16_+_42464613 | 0.04 |
ENSDART00000162454
|
si:ch211-215k15.4
|
si:ch211-215k15.4 |
| chr14_-_38872536 | 0.04 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr7_+_52178109 | 0.04 |
ENSDART00000174099
|
katnb1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr21_-_2955181 | 0.04 |
ENSDART00000158991
ENSDART00000174052 |
znf971
|
zinc finger protein 971 |
| chr2_+_37134281 | 0.04 |
ENSDART00000020135
|
pex19
|
peroxisomal biogenesis factor 19 |
| chr12_+_18458502 | 0.03 |
ENSDART00000108745
|
rnf151
|
ring finger protein 151 |
| chr19_-_32849036 | 0.03 |
ENSDART00000131518
|
si:dkey-284p5.3
|
si:dkey-284p5.3 |
| chr3_+_53240562 | 0.03 |
ENSDART00000031234
|
stxbp2
|
syntaxin binding protein 2 |
| chr22_+_24770744 | 0.03 |
ENSDART00000142882
|
si:rp71-23d18.4
|
si:rp71-23d18.4 |
| chr5_+_13385837 | 0.03 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr16_-_17056630 | 0.03 |
ENSDART00000138715
|
plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr6_-_40885496 | 0.03 |
ENSDART00000189857
|
sirt4
|
sirtuin 4 |
| chr5_-_20446082 | 0.03 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr13_-_8279533 | 0.03 |
ENSDART00000109666
|
si:ch211-250c4.5
|
si:ch211-250c4.5 |
| chr4_-_77380592 | 0.03 |
ENSDART00000163149
|
slco1e1
|
solute carrier organic anion transporter family, member 1E1 |
| chr19_+_22727940 | 0.02 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
| chr7_-_51953807 | 0.02 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr16_-_40373836 | 0.02 |
ENSDART00000134498
|
si:dkey-242e21.3
|
si:dkey-242e21.3 |
| chr4_+_59748607 | 0.02 |
ENSDART00000108499
|
znf1068
|
zinc finger protein 1068 |
| chr11_-_44498975 | 0.02 |
ENSDART00000173066
ENSDART00000189976 |
si:ch1073-365p7.2
|
si:ch1073-365p7.2 |
| chr2_-_51500957 | 0.02 |
ENSDART00000172481
|
pigrl3.5
|
polymeric immunoglobulin receptor-like 3.5 |
| chr15_-_37779978 | 0.02 |
ENSDART00000157202
|
si:dkey-42l23.3
|
si:dkey-42l23.3 |
| chr5_+_37068223 | 0.02 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr5_-_36674139 | 0.02 |
ENSDART00000188100
|
BX005407.1
|
|
| chr20_+_38322444 | 0.02 |
ENSDART00000161741
ENSDART00000132241 ENSDART00000148936 ENSDART00000149623 |
stum
|
stum, mechanosensory transduction mediator homolog |
| chr12_-_4916969 | 0.02 |
ENSDART00000161680
|
ifnphi4
|
interferon phi 4 |
| chr4_-_61650771 | 0.02 |
ENSDART00000168593
|
si:dkey-26i24.1
|
si:dkey-26i24.1 |
| chr6_+_3516926 | 0.01 |
ENSDART00000151607
|
faah
|
fatty acid amide hydrolase |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox14+sox14-1+sox21a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 1.8 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 2.0 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 0.7 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.2 | 0.5 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 1.0 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.1 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 9.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.3 | GO:0051701 | interaction with host(GO:0051701) |
| 0.1 | 0.3 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.1 | 0.6 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.2 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.2 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.2 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.2 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.2 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.2 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.0 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 2.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.7 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 9.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |