Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
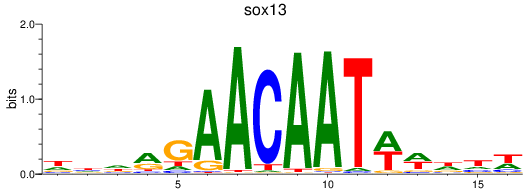
Results for sox13
Z-value: 0.81
Transcription factors associated with sox13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox13
|
ENSDARG00000030297 | SRY-box transcription factor 13 |
|
sox13
|
ENSDARG00000112638 | SRY-box transcription factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox13 | dr11_v1_chr11_+_37803773_37803773 | -0.67 | 4.8e-02 | Click! |
Activity profile of sox13 motif
Sorted Z-values of sox13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_24448278 | 1.87 |
ENSDART00000057584
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr23_+_31815423 | 1.38 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr5_+_32206378 | 1.18 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr9_-_22339582 | 1.06 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr7_+_39444843 | 1.05 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr7_-_8374950 | 1.01 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr3_+_28939759 | 0.97 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr12_-_20340543 | 0.97 |
ENSDART00000055623
|
hbbe3
|
hemoglobin beta embryonic-3 |
| chr23_-_42232124 | 0.94 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr6_-_42003780 | 0.89 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr6_-_58764672 | 0.84 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr3_-_61205711 | 0.81 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr14_-_25599002 | 0.81 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr5_-_67471375 | 0.74 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr5_-_32274383 | 0.73 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr21_+_26390549 | 0.73 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr12_+_15002757 | 0.71 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr21_-_25741411 | 0.70 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr22_-_15602760 | 0.68 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr8_-_49431939 | 0.67 |
ENSDART00000011453
ENSDART00000088240 ENSDART00000114173 |
sypb
|
synaptophysin b |
| chr14_-_12071679 | 0.65 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr25_+_31868268 | 0.65 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr7_-_51757085 | 0.65 |
ENSDART00000167747
|
leap2
|
liver-expressed antimicrobial peptide 2 |
| chr1_-_19079957 | 0.65 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr21_-_25741096 | 0.64 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr12_+_47663419 | 0.64 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr19_-_3931917 | 0.64 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr9_-_22318511 | 0.63 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr20_+_10538025 | 0.63 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr25_+_20215964 | 0.63 |
ENSDART00000139235
|
tnnt2d
|
troponin T2d, cardiac |
| chr20_-_14054083 | 0.62 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr10_-_7785930 | 0.61 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr2_-_16565690 | 0.61 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr20_-_40370736 | 0.61 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr22_-_17606575 | 0.59 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr19_+_1835940 | 0.58 |
ENSDART00000167372
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr11_-_29657947 | 0.57 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr23_-_22650992 | 0.57 |
ENSDART00000079007
|
ca6
|
carbonic anhydrase VI |
| chr6_+_12853655 | 0.57 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr25_+_20216159 | 0.57 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr14_+_34486629 | 0.57 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr5_-_31875645 | 0.56 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr6_+_12968101 | 0.55 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr3_-_37759969 | 0.55 |
ENSDART00000151105
ENSDART00000151208 |
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr23_-_22650668 | 0.55 |
ENSDART00000132733
|
ca6
|
carbonic anhydrase VI |
| chr3_-_30218504 | 0.54 |
ENSDART00000103487
|
zgc:195001
|
zgc:195001 |
| chr21_-_11834452 | 0.54 |
ENSDART00000081652
|
rfesd
|
Rieske (Fe-S) domain containing |
| chr20_-_29474859 | 0.53 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr16_-_55028740 | 0.52 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr20_-_29475172 | 0.52 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr12_+_20352400 | 0.52 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr21_+_19635486 | 0.51 |
ENSDART00000185736
|
fgf10a
|
fibroblast growth factor 10a |
| chr23_+_36771593 | 0.51 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr3_-_37759373 | 0.50 |
ENSDART00000150962
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr3_+_32689707 | 0.50 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr6_-_39275793 | 0.50 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr19_-_45960191 | 0.49 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr17_-_6382392 | 0.49 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr25_-_3830272 | 0.48 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr1_+_44906517 | 0.48 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr20_+_26538137 | 0.47 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr10_-_24391030 | 0.47 |
ENSDART00000180013
ENSDART00000177914 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr4_-_10599062 | 0.46 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr2_+_16780643 | 0.46 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr1_-_58064738 | 0.45 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr13_-_23074454 | 0.45 |
ENSDART00000065652
|
srgn
|
serglycin |
| chr20_-_40451115 | 0.45 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr7_+_58686860 | 0.45 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr9_-_42696408 | 0.44 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr22_+_3153876 | 0.44 |
ENSDART00000163327
|
rpl36
|
ribosomal protein L36 |
| chr9_+_17309195 | 0.44 |
ENSDART00000048548
|
scel
|
sciellin |
| chr3_+_26081343 | 0.44 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr24_+_22485710 | 0.43 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr5_-_36948586 | 0.43 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr7_-_8490886 | 0.43 |
ENSDART00000159012
|
jac6
|
jacalin 6 |
| chr15_-_2640966 | 0.43 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr16_-_17072440 | 0.42 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr7_+_25033924 | 0.41 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr12_-_35988586 | 0.41 |
ENSDART00000157746
|
pde6gb
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog b |
| chr22_-_15602593 | 0.41 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr22_+_38192568 | 0.41 |
ENSDART00000126323
|
cp
|
ceruloplasmin |
| chr11_-_1509773 | 0.40 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr4_+_57101749 | 0.40 |
ENSDART00000135121
|
si:ch211-238e22.4
|
si:ch211-238e22.4 |
| chr4_-_16330368 | 0.40 |
ENSDART00000128932
|
epyc
|
epiphycan |
| chr13_-_31470439 | 0.40 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr3_+_12764894 | 0.40 |
ENSDART00000166071
|
cyp2k16
|
cytochrome P450, family 2, subfamily K, polypeptide16 |
| chr10_-_11012000 | 0.39 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr15_+_28685625 | 0.39 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr3_+_32492467 | 0.39 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr15_-_21155641 | 0.39 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr2_-_44183613 | 0.38 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr18_+_30028637 | 0.38 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr21_-_35853245 | 0.37 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr7_-_28696556 | 0.37 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr4_+_25704579 | 0.37 |
ENSDART00000066945
|
lamb1b
|
laminin, beta 1b |
| chr21_-_8513192 | 0.37 |
ENSDART00000084378
|
crb2a
|
crumbs family member 2a |
| chr24_+_17269849 | 0.36 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr21_+_35215810 | 0.36 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr20_+_54079341 | 0.36 |
ENSDART00000060444
|
rps29
|
ribosomal protein S29 |
| chr18_+_45781516 | 0.36 |
ENSDART00000168840
|
rpl35a
|
ribosomal protein L35a |
| chr15_-_20233105 | 0.36 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr20_-_27225876 | 0.35 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr24_-_39567 | 0.35 |
ENSDART00000055488
|
cdh7b
|
cadherin 7b |
| chr24_-_26302375 | 0.35 |
ENSDART00000130696
|
cops9
|
COP9 signalosome subunit 9 |
| chr12_+_30586599 | 0.35 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr23_-_7799184 | 0.34 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr15_+_19797918 | 0.34 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr13_-_23074238 | 0.34 |
ENSDART00000143210
|
srgn
|
serglycin |
| chr3_-_30158395 | 0.34 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr23_+_22200467 | 0.34 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr14_-_4273396 | 0.33 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr25_-_19433244 | 0.33 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr1_+_54908895 | 0.33 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr22_-_15957041 | 0.33 |
ENSDART00000149236
ENSDART00000187500 ENSDART00000176304 ENSDART00000080047 ENSDART00000190068 |
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr5_+_27434601 | 0.33 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr13_+_28417297 | 0.32 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr11_-_7355995 | 0.32 |
ENSDART00000156595
|
si:dkey-197i20.6
|
si:dkey-197i20.6 |
| chr6_-_40195510 | 0.32 |
ENSDART00000156156
|
col7a1
|
collagen, type VII, alpha 1 |
| chr16_-_23346095 | 0.32 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr2_-_44183451 | 0.32 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr24_+_17270129 | 0.31 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr9_-_21498880 | 0.31 |
ENSDART00000131634
|
aff3
|
AF4/FMR2 family, member 3 |
| chr8_-_32497815 | 0.31 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr7_+_65673885 | 0.31 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr7_+_58699718 | 0.31 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr20_+_34845997 | 0.31 |
ENSDART00000035612
|
emilin1a
|
elastin microfibril interfacer 1a |
| chr6_-_39765546 | 0.31 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr5_-_64883082 | 0.31 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr4_+_69150355 | 0.30 |
ENSDART00000172364
ENSDART00000184374 |
si:ch211-209j12.2
|
si:ch211-209j12.2 |
| chr23_+_42819221 | 0.30 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr19_+_47311020 | 0.30 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr2_+_20406399 | 0.30 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr12_+_22258962 | 0.30 |
ENSDART00000131175
|
wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr1_-_21599219 | 0.29 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr16_-_42014272 | 0.29 |
ENSDART00000180488
|
etv2
|
ets variant 2 |
| chr19_-_10881486 | 0.29 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr21_+_5589923 | 0.29 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr10_+_35439952 | 0.29 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr16_+_42772678 | 0.29 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr10_+_21559605 | 0.29 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr14_-_11456724 | 0.29 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr23_+_31107685 | 0.29 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr19_+_4990320 | 0.29 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr8_-_32497581 | 0.29 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr23_-_15216654 | 0.29 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr4_+_17279966 | 0.29 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr19_-_47527093 | 0.28 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr4_+_16794418 | 0.28 |
ENSDART00000079429
|
spx
|
spexin hormone |
| chr19_+_47311869 | 0.28 |
ENSDART00000136647
|
ext1c
|
exostoses (multiple) 1c |
| chr25_-_35360096 | 0.28 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
| chr16_+_5184402 | 0.28 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr3_+_29445473 | 0.28 |
ENSDART00000020381
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr17_+_4207278 | 0.27 |
ENSDART00000193612
|
CU929304.1
|
|
| chr7_+_60079302 | 0.27 |
ENSDART00000051524
|
etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr11_-_23458792 | 0.27 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr16_+_13855039 | 0.27 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr21_-_5879897 | 0.27 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr24_-_31306724 | 0.27 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr2_+_47905735 | 0.27 |
ENSDART00000159701
|
ftr23
|
finTRIM family, member 23 |
| chr19_+_26718074 | 0.27 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr21_+_30937690 | 0.27 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr18_+_17583479 | 0.27 |
ENSDART00000186977
ENSDART00000010998 |
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr3_+_12835370 | 0.27 |
ENSDART00000166089
|
si:ch211-8c17.3
|
si:ch211-8c17.3 |
| chr11_-_29737088 | 0.27 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr24_+_39105051 | 0.26 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr5_-_35301800 | 0.26 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr14_-_33454595 | 0.26 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr7_-_35516251 | 0.26 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr13_+_4405282 | 0.26 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr14_+_36218072 | 0.26 |
ENSDART00000181864
|
pitx2
|
paired-like homeodomain 2 |
| chr4_+_31293362 | 0.26 |
ENSDART00000169781
|
si:dkey-57l19.1
|
si:dkey-57l19.1 |
| chr14_-_2202652 | 0.25 |
ENSDART00000171316
|
si:dkey-262j3.7
|
si:dkey-262j3.7 |
| chr25_-_12923482 | 0.25 |
ENSDART00000161754
|
CR450808.1
|
|
| chr3_+_60716904 | 0.25 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr12_-_20584413 | 0.25 |
ENSDART00000170923
|
FP885542.2
|
|
| chr16_+_31802203 | 0.25 |
ENSDART00000058739
ENSDART00000110834 |
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr15_-_19724932 | 0.24 |
ENSDART00000152345
|
sytl2b
|
synaptotagmin-like 2b |
| chr2_-_9646857 | 0.24 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr3_-_28828242 | 0.24 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr7_+_57795974 | 0.24 |
ENSDART00000148369
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr13_-_18863680 | 0.24 |
ENSDART00000109277
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr19_-_32150078 | 0.24 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr13_-_40398556 | 0.24 |
ENSDART00000191921
|
nkx3.3
|
NK3 homeobox 3 |
| chr8_-_50259448 | 0.24 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr16_+_37582872 | 0.24 |
ENSDART00000169331
|
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr5_+_46196147 | 0.24 |
ENSDART00000084112
|
f2r
|
coagulation factor II (thrombin) receptor |
| chr8_+_3405612 | 0.24 |
ENSDART00000163437
|
zgc:112433
|
zgc:112433 |
| chr7_+_20467549 | 0.23 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr23_-_15330168 | 0.23 |
ENSDART00000035865
ENSDART00000143635 |
sulf2b
|
sulfatase 2b |
| chr8_-_23776399 | 0.23 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr17_-_31695217 | 0.23 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr8_+_32755688 | 0.23 |
ENSDART00000137897
|
hmcn2
|
hemicentin 2 |
| chr4_-_18315912 | 0.23 |
ENSDART00000135735
|
plxnc1
|
plexin C1 |
| chr12_+_25097754 | 0.23 |
ENSDART00000050070
|
pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr19_-_41405187 | 0.23 |
ENSDART00000038038
ENSDART00000172350 |
sem1
|
SEM1, 26S proteasome complex subunit |
| chr21_-_40676224 | 0.23 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr21_+_19062124 | 0.22 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr22_+_10651726 | 0.22 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr8_-_16499508 | 0.22 |
ENSDART00000108659
|
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr23_+_45512825 | 0.22 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.2 | 0.8 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.2 | 1.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.6 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.2 | 0.6 | GO:0015840 | urea transport(GO:0015840) |
| 0.1 | 0.4 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.4 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.5 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.4 | GO:0090076 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.3 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 0.3 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.1 | 0.8 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.2 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 2.1 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.1 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.5 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.2 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.4 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.6 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.1 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.1 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 2.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 0.3 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.6 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 1.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.6 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.0 | 0.8 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.2 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.1 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.2 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 | 0.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.1 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.4 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.3 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.3 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.1 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0030072 | peptide secretion(GO:0002790) peptide hormone secretion(GO:0030072) insulin secretion(GO:0030073) |
| 0.0 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 1.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.3 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 1.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.6 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0042706 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 1.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.6 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 1.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.7 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.8 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 2.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.8 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.6 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 1.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.2 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |