Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
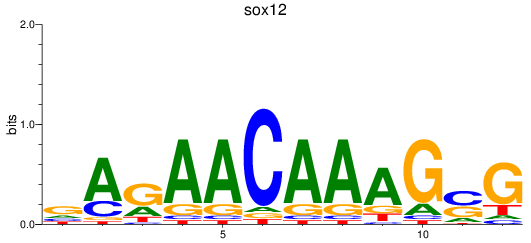
Results for sox12
Z-value: 1.13
Transcription factors associated with sox12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox12
|
ENSDARG00000025847 | SRY-box transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox12 | dr11_v1_chr11_-_24191928_24191928 | -0.64 | 6.1e-02 | Click! |
Activity profile of sox12 motif
Sorted Z-values of sox12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_35445462 | 2.55 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr20_-_14114078 | 2.00 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr17_-_26507289 | 1.94 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr16_-_41762983 | 1.91 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr7_-_24520866 | 1.87 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr5_-_28149767 | 1.77 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr5_-_38384755 | 1.71 |
ENSDART00000188573
ENSDART00000051233 |
mink1
|
misshapen-like kinase 1 |
| chr22_-_31517300 | 1.60 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr4_-_2945306 | 1.53 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr18_-_11595567 | 1.48 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr17_+_43595692 | 1.47 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr4_+_11723852 | 1.42 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr22_+_34784075 | 1.40 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr5_+_40835601 | 1.39 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr3_-_13955878 | 1.39 |
ENSDART00000166804
|
gcdhb
|
glutaryl-CoA dehydrogenase b |
| chr17_+_7534180 | 1.38 |
ENSDART00000187512
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr10_+_10972795 | 1.32 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr8_+_54135642 | 1.30 |
ENSDART00000170712
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr5_+_45138934 | 1.30 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_24520518 | 1.29 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr17_+_7534365 | 1.28 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr10_-_32660716 | 1.27 |
ENSDART00000063544
ENSDART00000141013 |
atg101
|
autophagy related 101 |
| chr20_-_23876291 | 1.25 |
ENSDART00000043316
|
katna1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr21_+_13244450 | 1.25 |
ENSDART00000146062
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr5_-_38384289 | 1.23 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr18_+_8912113 | 1.23 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr6_-_13022166 | 1.22 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr18_+_8912536 | 1.21 |
ENSDART00000134827
ENSDART00000061904 |
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr7_-_735118 | 1.21 |
ENSDART00000126424
|
MAP3K11
|
si:cabz01078036.1 |
| chr25_+_33063762 | 1.19 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr8_-_38201415 | 1.17 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr20_+_23083800 | 1.15 |
ENSDART00000132093
|
usp46
|
ubiquitin specific peptidase 46 |
| chr5_+_45139196 | 1.12 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_43456025 | 1.11 |
ENSDART00000001092
ENSDART00000140618 |
ncor2
|
nuclear receptor corepressor 2 |
| chr11_+_24820542 | 1.08 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr12_-_17863467 | 1.08 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr4_-_14531687 | 1.08 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr11_+_31864921 | 1.08 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr20_+_4221978 | 1.03 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr10_+_3153973 | 1.03 |
ENSDART00000183223
|
hic2
|
hypermethylated in cancer 2 |
| chr7_+_24573721 | 1.02 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr17_-_7218481 | 1.01 |
ENSDART00000181967
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr5_-_37875636 | 0.99 |
ENSDART00000184674
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr3_-_60856157 | 0.98 |
ENSDART00000053502
|
CABZ01087513.1
|
|
| chr7_-_24995631 | 0.98 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr20_+_1385674 | 0.96 |
ENSDART00000145981
|
mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr5_+_3891485 | 0.95 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr25_+_22017182 | 0.95 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr5_-_24231139 | 0.93 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr1_+_47331644 | 0.92 |
ENSDART00000053284
|
bcl9
|
B cell CLL/lymphoma 9 |
| chr16_-_32975951 | 0.92 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr1_-_38361496 | 0.91 |
ENSDART00000015323
|
fbxo8
|
F-box protein 8 |
| chr22_-_9861531 | 0.90 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr1_+_40613297 | 0.90 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr20_-_2355357 | 0.89 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr15_-_35930070 | 0.89 |
ENSDART00000076229
|
irs1
|
insulin receptor substrate 1 |
| chr11_+_6115621 | 0.89 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr1_+_45839927 | 0.88 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr15_-_28587147 | 0.87 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr10_+_5268054 | 0.86 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr6_-_41135215 | 0.85 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr2_-_31833347 | 0.84 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr15_+_31911989 | 0.82 |
ENSDART00000111472
|
brca2
|
breast cancer 2, early onset |
| chr8_+_16990120 | 0.79 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr17_-_23709347 | 0.77 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr20_+_4222357 | 0.77 |
ENSDART00000188331
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr18_+_8912710 | 0.76 |
ENSDART00000142866
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr3_-_1146497 | 0.76 |
ENSDART00000149061
|
si:ch73-211l13.2
|
si:ch73-211l13.2 |
| chr7_+_24729558 | 0.76 |
ENSDART00000111542
ENSDART00000170100 |
shroom4
|
shroom family member 4 |
| chr11_-_21267486 | 0.75 |
ENSDART00000128681
|
DYRK3
|
zgc:172180 |
| chr1_-_52790724 | 0.75 |
ENSDART00000139577
ENSDART00000100937 |
patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr22_-_9860792 | 0.74 |
ENSDART00000155908
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr24_-_41267184 | 0.74 |
ENSDART00000063504
|
xylb
|
xylulokinase homolog (H. influenzae) |
| chr15_-_1484795 | 0.73 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr24_-_15263142 | 0.71 |
ENSDART00000183176
ENSDART00000006930 |
rttn
|
rotatin |
| chr3_-_33175583 | 0.71 |
ENSDART00000126022
|
rarab
|
retinoic acid receptor, alpha b |
| chr1_+_46509176 | 0.70 |
ENSDART00000166028
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr11_-_16394971 | 0.70 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr6_+_18367388 | 0.70 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr16_+_23796612 | 0.69 |
ENSDART00000131698
|
rab13
|
RAB13, member RAS oncogene family |
| chr21_+_31150438 | 0.69 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr13_-_33317323 | 0.67 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr2_+_4303125 | 0.65 |
ENSDART00000161211
ENSDART00000162269 |
cables1
|
Cdk5 and Abl enzyme substrate 1 |
| chr15_-_16946124 | 0.64 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr16_-_16237844 | 0.61 |
ENSDART00000168747
ENSDART00000111912 |
rbm12b
|
RNA binding motif protein 12B |
| chr2_-_24069331 | 0.61 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr3_-_35865040 | 0.61 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr18_+_22138924 | 0.60 |
ENSDART00000183961
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr8_+_26859639 | 0.60 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr18_+_6479963 | 0.59 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr5_+_58455488 | 0.59 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr13_-_12667220 | 0.59 |
ENSDART00000079594
|
fam241a
|
family with sequence similarity 241 member A |
| chr11_+_1602916 | 0.58 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr7_+_25003313 | 0.55 |
ENSDART00000131935
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr1_+_594736 | 0.55 |
ENSDART00000166731
|
jam2a
|
junctional adhesion molecule 2a |
| chr9_-_16140860 | 0.54 |
ENSDART00000142974
|
myo1b
|
myosin IB |
| chr10_+_8554929 | 0.52 |
ENSDART00000190849
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr6_+_36942966 | 0.51 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr23_-_36449111 | 0.51 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr22_-_784110 | 0.51 |
ENSDART00000061775
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr1_-_26023678 | 0.50 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr16_+_32082547 | 0.50 |
ENSDART00000190122
|
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr16_+_32082359 | 0.50 |
ENSDART00000140794
ENSDART00000137029 |
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr3_-_33437796 | 0.50 |
ENSDART00000075499
|
si:dkey-283b1.7
|
si:dkey-283b1.7 |
| chr14_-_48033073 | 0.49 |
ENSDART00000193115
ENSDART00000169300 ENSDART00000188036 ENSDART00000183432 ENSDART00000180973 |
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr22_+_21618121 | 0.49 |
ENSDART00000133939
|
tle2a
|
transducin like enhancer of split 2a |
| chr7_+_29115890 | 0.48 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr13_+_9559461 | 0.48 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr16_+_25343934 | 0.45 |
ENSDART00000109710
|
zfat
|
zinc finger and AT hook domain containing |
| chr9_+_37329036 | 0.45 |
ENSDART00000131756
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr14_+_48960078 | 0.43 |
ENSDART00000165514
|
mif
|
macrophage migration inhibitory factor |
| chr2_-_24068848 | 0.43 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr8_-_2153147 | 0.41 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr23_+_9268083 | 0.41 |
ENSDART00000055054
|
acss2
|
acyl-CoA synthetase short chain family member 2 |
| chr10_-_42131408 | 0.40 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr23_-_7755373 | 0.39 |
ENSDART00000162868
|
PCMTD2 (1 of many)
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr17_+_30843881 | 0.38 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr11_+_37768298 | 0.37 |
ENSDART00000166886
|
sox13
|
SRY (sex determining region Y)-box 13 |
| chr12_-_22400999 | 0.37 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr16_+_11242443 | 0.35 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr3_+_61523035 | 0.34 |
ENSDART00000106530
|
znf1004
|
zinc finger protein 1004 |
| chr23_+_30707837 | 0.34 |
ENSDART00000016096
|
dnajc11a
|
DnaJ (Hsp40) homolog, subfamily C, member 11a |
| chr17_-_15229787 | 0.33 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr8_+_35356944 | 0.31 |
ENSDART00000143243
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr16_-_28091597 | 0.31 |
ENSDART00000166595
|
CR626941.1
|
|
| chr10_-_25204034 | 0.30 |
ENSDART00000161100
|
FAT3 (1 of many)
|
si:ch211-214k5.6 |
| chr1_+_41487339 | 0.29 |
ENSDART00000142508
ENSDART00000114449 |
si:ch211-89o9.6
|
si:ch211-89o9.6 |
| chr5_+_26221011 | 0.28 |
ENSDART00000007587
|
gtf2h2
|
general transcription factor IIH, polypeptide 2 |
| chr7_-_28565230 | 0.25 |
ENSDART00000028887
|
tmem9b
|
TMEM9 domain family, member B |
| chr8_-_52229462 | 0.25 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr19_-_38830582 | 0.24 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr16_-_9897769 | 0.22 |
ENSDART00000187044
|
ncalda
|
neurocalcin delta a |
| chr15_+_32419303 | 0.22 |
ENSDART00000162663
|
si:dkey-285b23.3
|
si:dkey-285b23.3 |
| chr13_+_35765317 | 0.19 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr8_-_13419049 | 0.18 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr8_-_13471916 | 0.18 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr16_-_9897613 | 0.18 |
ENSDART00000104058
|
ncalda
|
neurocalcin delta a |
| chr9_+_24920677 | 0.17 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr10_-_35321625 | 0.16 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr24_-_37698796 | 0.16 |
ENSDART00000172178
|
si:ch211-231f6.6
|
si:ch211-231f6.6 |
| chr19_+_20708460 | 0.16 |
ENSDART00000034124
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr18_+_45504362 | 0.15 |
ENSDART00000140089
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr6_+_33897624 | 0.13 |
ENSDART00000166706
|
ccdc17
|
coiled-coil domain containing 17 |
| chr17_-_21057617 | 0.13 |
ENSDART00000148095
ENSDART00000048853 |
ube2d1a
|
ubiquitin-conjugating enzyme E2D 1a |
| chr2_-_16217344 | 0.13 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr17_+_45654724 | 0.13 |
ENSDART00000098952
|
zgc:162184
|
zgc:162184 |
| chr3_-_34027178 | 0.12 |
ENSDART00000170201
ENSDART00000151408 |
ighv1-4
ighv14-1
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 14-1 |
| chr12_-_29301022 | 0.12 |
ENSDART00000187826
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr5_+_42124706 | 0.11 |
ENSDART00000020044
ENSDART00000156372 |
shpk
|
sedoheptulokinase |
| chr17_-_25303486 | 0.11 |
ENSDART00000162235
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr15_+_26933196 | 0.11 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr8_+_9149436 | 0.11 |
ENSDART00000064090
|
pnck
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr7_+_49078890 | 0.09 |
ENSDART00000067857
|
b4galnt4b
|
beta-1,4-N-acetyl-galactosaminyl transferase 4b |
| chr3_-_40284744 | 0.09 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr6_+_27667359 | 0.08 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr8_+_30600386 | 0.07 |
ENSDART00000164976
|
specc1la
|
sperm antigen with calponin homology and coiled-coil domains 1-like a |
| chr9_-_53817143 | 0.07 |
ENSDART00000148764
ENSDART00000075537 |
ednrbb
|
endothelin receptor type Bb |
| chr15_+_37545855 | 0.07 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr1_-_26071535 | 0.07 |
ENSDART00000185187
|
pdcd4a
|
programmed cell death 4a |
| chr3_-_16110351 | 0.06 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr4_-_77380592 | 0.06 |
ENSDART00000163149
|
slco1e1
|
solute carrier organic anion transporter family, member 1E1 |
| chr14_-_2933185 | 0.06 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr7_-_30217000 | 0.06 |
ENSDART00000173646
|
znf710b
|
zinc finger protein 710b |
| chr6_-_55347616 | 0.05 |
ENSDART00000172081
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr4_-_2525916 | 0.05 |
ENSDART00000134123
ENSDART00000132581 ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr25_-_31171948 | 0.05 |
ENSDART00000184275
|
FQ976913.1
|
|
| chr17_-_49443399 | 0.04 |
ENSDART00000155293
|
cga
|
glycoprotein hormones, alpha polypeptide |
| chr17_-_7436766 | 0.04 |
ENSDART00000162002
|
grm1b
|
glutamate receptor, metabotropic 1b |
| chr8_-_19313510 | 0.03 |
ENSDART00000164780
ENSDART00000137133 |
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr3_+_21669545 | 0.02 |
ENSDART00000156527
|
crhr1
|
corticotropin releasing hormone receptor 1 |
| chr3_+_3545825 | 0.02 |
ENSDART00000109060
|
CR589947.1
|
|
| chr9_+_15893093 | 0.02 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr25_+_25124684 | 0.02 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr6_+_46406565 | 0.00 |
ENSDART00000168440
ENSDART00000131203 ENSDART00000138567 ENSDART00000132845 |
pbrm1l
|
polybromo 1, like |
| chr3_-_16110100 | 0.00 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.4 | 2.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 0.8 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.7 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 2.6 | GO:0030719 | P granule organization(GO:0030719) |
| 0.2 | 0.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 1.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 0.7 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 1.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 1.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.9 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 1.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.9 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.6 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 2.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.4 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.3 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.4 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.1 | 0.7 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.8 | GO:0008585 | sex determination(GO:0007530) female gonad development(GO:0008585) |
| 0.1 | 0.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.0 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.2 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 1.1 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 1.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.9 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.7 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 0.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.0 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 2.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.3 | 0.8 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.9 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.6 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 0.9 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.6 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.4 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 2.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.1 | 0.4 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 2.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.0 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.1 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.4 | GO:0070122 | Lys63-specific deubiquitinase activity(GO:0061578) isopeptidase activity(GO:0070122) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.1 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.9 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.9 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |