Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
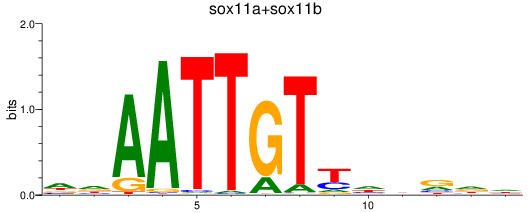
Results for sox11a+sox11b
Z-value: 0.62
Transcription factors associated with sox11a+sox11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox11a
|
ENSDARG00000077811 | SRY-box transcription factor 11a |
|
sox11b
|
ENSDARG00000095743 | SRY-box transcription factor 11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox11a | dr11_v1_chr17_-_35881841_35881841 | 0.54 | 1.4e-01 | Click! |
| sox11b | dr11_v1_chr20_-_30035326_30035326 | 0.39 | 3.0e-01 | Click! |
Activity profile of sox11a+sox11b motif
Sorted Z-values of sox11a+sox11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22240052 | 1.15 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr3_-_61205711 | 0.88 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr12_-_20340543 | 0.78 |
ENSDART00000055623
|
hbbe3
|
hemoglobin beta embryonic-3 |
| chr22_-_26595027 | 0.77 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr5_-_32308526 | 0.77 |
ENSDART00000193758
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr9_+_32978302 | 0.72 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr21_-_41305748 | 0.72 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr21_+_26389391 | 0.69 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr2_+_16780643 | 0.57 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr5_+_26795465 | 0.51 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr7_-_8490886 | 0.51 |
ENSDART00000159012
|
jac6
|
jacalin 6 |
| chr23_-_22650992 | 0.49 |
ENSDART00000079007
|
ca6
|
carbonic anhydrase VI |
| chr16_-_24598042 | 0.48 |
ENSDART00000156407
ENSDART00000154072 |
si:dkey-56f14.4
|
si:dkey-56f14.4 |
| chr15_-_15357178 | 0.47 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr19_+_26718074 | 0.47 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr7_-_58729894 | 0.47 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr16_-_23346095 | 0.46 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr20_+_37295006 | 0.46 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr1_-_14234076 | 0.45 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr18_+_20494413 | 0.44 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr4_+_14957360 | 0.44 |
ENSDART00000002770
ENSDART00000111882 ENSDART00000148292 |
tspan33a
|
tetraspanin 33a |
| chr1_-_20928772 | 0.43 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr23_+_31815423 | 0.42 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr13_+_24755049 | 0.42 |
ENSDART00000134102
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr22_-_17606575 | 0.41 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr6_-_40195510 | 0.40 |
ENSDART00000156156
|
col7a1
|
collagen, type VII, alpha 1 |
| chr11_+_30513656 | 0.38 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr10_-_28761454 | 0.37 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr5_-_67911111 | 0.37 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr9_-_1949915 | 0.36 |
ENSDART00000190712
|
hoxd3a
|
homeobox D3a |
| chr10_+_21650828 | 0.33 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr20_-_16171297 | 0.33 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr19_-_47456787 | 0.32 |
ENSDART00000168792
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr14_+_4151379 | 0.32 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr14_-_26704829 | 0.31 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr9_+_24008879 | 0.31 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr19_-_5369486 | 0.30 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr23_+_23022790 | 0.30 |
ENSDART00000189843
|
samd11
|
sterile alpha motif domain containing 11 |
| chr20_-_26039841 | 0.30 |
ENSDART00000179929
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr23_-_5685023 | 0.30 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr13_+_13693722 | 0.27 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr3_-_12930217 | 0.26 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr21_-_41025340 | 0.26 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr2_+_24177006 | 0.25 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr5_+_69716458 | 0.25 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr12_+_22576404 | 0.23 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr22_+_6674992 | 0.22 |
ENSDART00000144054
|
si:ch211-209l18.2
|
si:ch211-209l18.2 |
| chr17_+_33340675 | 0.22 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr20_-_47731768 | 0.22 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr8_-_38105053 | 0.22 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr22_+_3156386 | 0.22 |
ENSDART00000161212
|
rpl36
|
ribosomal protein L36 |
| chr11_-_30158191 | 0.21 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr7_-_8515550 | 0.21 |
ENSDART00000111896
|
jac9
|
jacalin 9 |
| chr16_-_17072440 | 0.20 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr11_+_41243094 | 0.20 |
ENSDART00000191782
ENSDART00000169817 ENSDART00000162157 |
pax7a
|
paired box 7a |
| chr7_-_35314347 | 0.19 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr3_+_22375596 | 0.19 |
ENSDART00000188243
ENSDART00000181506 |
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr5_-_35301800 | 0.19 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr1_-_35695614 | 0.19 |
ENSDART00000133813
ENSDART00000145291 ENSDART00000176451 |
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr1_-_36152131 | 0.19 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr12_+_47663419 | 0.19 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr17_-_48915427 | 0.19 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr20_+_44311448 | 0.18 |
ENSDART00000114660
|
opn8b
|
opsin 8, group member b |
| chr16_-_22006996 | 0.18 |
ENSDART00000116114
|
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr2_-_40135942 | 0.18 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr19_-_9522548 | 0.18 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr23_-_31403668 | 0.18 |
ENSDART00000147498
|
CR759830.1
|
|
| chr12_+_14676349 | 0.18 |
ENSDART00000143401
|
becn1
|
beclin 1, autophagy related |
| chr1_-_19079957 | 0.17 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr8_-_14050758 | 0.17 |
ENSDART00000133922
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr6_+_42820228 | 0.17 |
ENSDART00000185413
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr19_+_2279051 | 0.16 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr9_-_18568927 | 0.16 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr2_+_9061885 | 0.16 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr1_+_16397063 | 0.15 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr20_+_26538137 | 0.15 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr22_-_6991514 | 0.15 |
ENSDART00000148233
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr6_+_4229360 | 0.15 |
ENSDART00000191347
ENSDART00000130642 |
FO082877.1
|
|
| chr10_+_29259882 | 0.15 |
ENSDART00000180606
|
sytl2a
|
synaptotagmin-like 2a |
| chr19_+_23919096 | 0.14 |
ENSDART00000090200
|
snapin
|
SNAP-associated protein |
| chr20_+_19793620 | 0.14 |
ENSDART00000136317
|
chrna2b
|
cholinergic receptor, nicotinic, alpha 2b (neuronal) |
| chr4_+_28374628 | 0.13 |
ENSDART00000076037
|
alg10
|
asparagine-linked glycosylation 10 |
| chr24_-_37680917 | 0.13 |
ENSDART00000131342
|
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr8_-_39978767 | 0.13 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr20_+_54383838 | 0.13 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr5_-_41841892 | 0.13 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr23_-_17657348 | 0.13 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr11_+_30282141 | 0.13 |
ENSDART00000122756
|
si:dkey-163f14.6
|
si:dkey-163f14.6 |
| chr23_+_40460333 | 0.12 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr5_-_57528943 | 0.12 |
ENSDART00000130320
|
pisd
|
phosphatidylserine decarboxylase |
| chr25_+_17920361 | 0.12 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr1_-_34713710 | 0.12 |
ENSDART00000141569
ENSDART00000102111 ENSDART00000122137 |
tpt1
|
tumor protein, translationally-controlled 1 |
| chr5_+_66353750 | 0.12 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr2_+_24177190 | 0.12 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr20_-_38836161 | 0.12 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr16_+_23087326 | 0.12 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr25_+_19485198 | 0.11 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr18_+_25653599 | 0.11 |
ENSDART00000007856
|
fkbp16
|
FK506 binding protein 16 |
| chr6_+_39812475 | 0.11 |
ENSDART00000067063
|
c1ql4b
|
complement component 1, q subcomponent-like 4b |
| chr2_-_37889111 | 0.11 |
ENSDART00000168939
ENSDART00000098529 |
mbl2
|
mannose binding lectin 2 |
| chr5_+_66433287 | 0.11 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr12_-_37734973 | 0.11 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr18_+_30028637 | 0.10 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr8_-_5220125 | 0.10 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr11_+_39672874 | 0.10 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr15_-_14467394 | 0.09 |
ENSDART00000191944
|
numbl
|
numb homolog (Drosophila)-like |
| chr17_-_24439672 | 0.09 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr2_-_1569250 | 0.09 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr11_+_23760470 | 0.09 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr3_+_26267725 | 0.09 |
ENSDART00000131288
|
adap2
|
ArfGAP with dual PH domains 2 |
| chr19_-_21832441 | 0.09 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr25_+_14087045 | 0.09 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr24_-_26302375 | 0.09 |
ENSDART00000130696
|
cops9
|
COP9 signalosome subunit 9 |
| chr20_+_50852356 | 0.09 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr7_-_40145097 | 0.08 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr1_-_22512063 | 0.08 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr3_+_18425814 | 0.08 |
ENSDART00000144222
|
gaa
|
glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) |
| chr6_-_50704689 | 0.08 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr13_-_11644806 | 0.08 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr20_+_33519435 | 0.07 |
ENSDART00000061829
|
drc1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr14_-_2187202 | 0.07 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr2_+_28672152 | 0.07 |
ENSDART00000157410
ENSDART00000169614 |
nadsyn1
|
NAD synthetase 1 |
| chr12_-_21834611 | 0.07 |
ENSDART00000179553
|
thrab
|
thyroid hormone receptor alpha b |
| chr13_+_35637048 | 0.07 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr2_-_30055432 | 0.07 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr11_-_16093018 | 0.07 |
ENSDART00000139309
ENSDART00000139819 |
SPATA1
|
si:dkey-205k8.5 |
| chr7_+_54642005 | 0.07 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr5_-_3118346 | 0.07 |
ENSDART00000167554
|
hsf5
|
heat shock transcription factor family member 5 |
| chr21_+_42930558 | 0.06 |
ENSDART00000135234
|
stk32a
|
serine/threonine kinase 32A |
| chr17_+_52524452 | 0.06 |
ENSDART00000016614
|
dlst
|
dihydrolipoamide S-succinyltransferase |
| chr1_-_35924495 | 0.06 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr13_-_25198025 | 0.06 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr6_+_54538948 | 0.06 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr21_-_40317035 | 0.06 |
ENSDART00000143648
ENSDART00000013359 |
or101-1
|
odorant receptor, family B, subfamily 101, member 1 |
| chr10_-_27009413 | 0.06 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr20_-_8304271 | 0.06 |
ENSDART00000179057
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr16_+_36768674 | 0.06 |
ENSDART00000169208
ENSDART00000180470 |
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr19_-_31402429 | 0.06 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr15_-_38177768 | 0.05 |
ENSDART00000152404
|
si:dkey-24p1.7
|
si:dkey-24p1.7 |
| chr13_-_1349922 | 0.05 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr19_-_17864213 | 0.05 |
ENSDART00000151043
|
ints8
|
integrator complex subunit 8 |
| chr6_-_23931442 | 0.05 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr1_-_23596391 | 0.05 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr18_-_25177230 | 0.05 |
ENSDART00000013363
|
slco3a1
|
solute carrier organic anion transporter family, member 3A1 |
| chr1_-_21297748 | 0.05 |
ENSDART00000142109
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr2_+_47906240 | 0.05 |
ENSDART00000122206
|
ftr23
|
finTRIM family, member 23 |
| chr8_-_50888806 | 0.04 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr18_-_2222128 | 0.04 |
ENSDART00000171402
|
pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr20_-_31067306 | 0.04 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr16_-_15988320 | 0.04 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr10_-_39321367 | 0.04 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr23_+_35918530 | 0.04 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr10_+_19026988 | 0.04 |
ENSDART00000137456
|
igsf9a
|
immunoglobulin superfamily, member 9a |
| chr5_+_37069605 | 0.04 |
ENSDART00000161977
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr7_-_22981796 | 0.04 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr10_+_38806719 | 0.04 |
ENSDART00000184416
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr22_-_35961365 | 0.04 |
ENSDART00000188434
|
LO016987.2
|
|
| chr3_+_19299309 | 0.03 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr21_+_5080789 | 0.03 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chrM_+_6425 | 0.03 |
ENSDART00000093606
|
mt-co1
|
cytochrome c oxidase I, mitochondrial |
| chr3_+_22270463 | 0.03 |
ENSDART00000104004
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr12_+_19205086 | 0.03 |
ENSDART00000186573
|
sh3bp1
|
SH3-domain binding protein 1 |
| chr22_-_8692305 | 0.03 |
ENSDART00000181602
|
CR450686.4
|
|
| chr1_+_46948010 | 0.03 |
ENSDART00000132843
|
si:dkey-22n8.2
|
si:dkey-22n8.2 |
| chr4_-_25859305 | 0.03 |
ENSDART00000159528
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr2_-_32643738 | 0.03 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr8_-_10045043 | 0.03 |
ENSDART00000081917
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr20_-_47188966 | 0.03 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr16_-_33001153 | 0.03 |
ENSDART00000147941
|
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr25_+_19041329 | 0.03 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr12_+_46696867 | 0.03 |
ENSDART00000152928
ENSDART00000153445 ENSDART00000123357 ENSDART00000152880 ENSDART00000123834 ENSDART00000174765 ENSDART00000189923 |
exoc7
|
exocyst complex component 7 |
| chr3_+_12828824 | 0.03 |
ENSDART00000160552
|
si:ch211-8c17.2
|
si:ch211-8c17.2 |
| chr23_-_6865946 | 0.02 |
ENSDART00000056426
|
ftr58
|
finTRIM family, member 58 |
| chr17_+_12658411 | 0.02 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr12_-_44010532 | 0.02 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr16_+_54263921 | 0.02 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr7_+_22981909 | 0.02 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr21_-_38153824 | 0.02 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr22_+_17606863 | 0.02 |
ENSDART00000035670
|
polr2eb
|
polymerase (RNA) II (DNA directed) polypeptide E, b |
| chr23_-_33640959 | 0.02 |
ENSDART00000187599
ENSDART00000189475 |
csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr16_-_21140097 | 0.02 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr20_+_50115335 | 0.02 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr1_+_26445615 | 0.02 |
ENSDART00000180810
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr11_-_41222971 | 0.01 |
ENSDART00000163609
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr4_-_18635005 | 0.01 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr13_-_40499296 | 0.01 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr17_-_47090440 | 0.01 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr23_+_11669109 | 0.01 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr13_-_45201300 | 0.01 |
ENSDART00000074750
ENSDART00000180265 |
runx3
|
runt-related transcription factor 3 |
| chr8_-_36140405 | 0.01 |
ENSDART00000182806
|
CT583723.2
|
|
| chr5_-_61431809 | 0.01 |
ENSDART00000082952
|
rcc1l
|
RCC1 like |
| chr15_-_20949692 | 0.01 |
ENSDART00000185548
|
tbcela
|
tubulin folding cofactor E-like a |
| chr1_-_54765262 | 0.01 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr15_+_16908085 | 0.01 |
ENSDART00000186870
|
ypel2b
|
yippee-like 2b |
| chr6_-_1768724 | 0.01 |
ENSDART00000162488
ENSDART00000163613 |
zgc:158417
|
zgc:158417 |
| chr5_-_66301142 | 0.01 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr2_+_904362 | 0.01 |
ENSDART00000024967
|
ripk1l
|
receptor (TNFRSF)-interacting serine-threonine kinase 1, like |
| chr9_+_38883388 | 0.01 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr23_-_24234371 | 0.01 |
ENSDART00000124539
|
ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr22_-_23545307 | 0.00 |
ENSDART00000166915
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox11a+sox11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.5 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.4 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.1 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.2 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.1 | GO:0042040 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |