Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
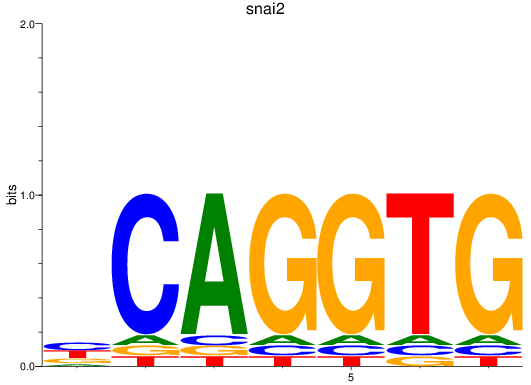
Results for snai2
Z-value: 1.03
Transcription factors associated with snai2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai2
|
ENSDARG00000040046 | snail family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai2 | dr11_v1_chr24_+_35387517_35387517 | -0.48 | 1.9e-01 | Click! |
Activity profile of snai2 motif
Sorted Z-values of snai2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_25331439 | 1.76 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr15_-_1590858 | 1.71 |
ENSDART00000081875
|
nnr
|
nanor |
| chr20_-_43743700 | 1.63 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr20_-_41992878 | 1.62 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr10_+_19596214 | 1.51 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr19_-_18313303 | 1.51 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr23_-_44574059 | 1.37 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr20_-_23426339 | 1.33 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_-_2596125 | 1.23 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr14_+_15155684 | 1.18 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr21_-_43666420 | 1.10 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr17_-_25330775 | 1.08 |
ENSDART00000154048
|
zpcx
|
zona pellucida protein C |
| chr3_+_1179601 | 0.98 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr16_+_52343905 | 0.95 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr15_+_784149 | 0.93 |
ENSDART00000155114
|
znf970
|
zinc finger protein 970 |
| chr7_-_51773166 | 0.92 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr17_+_1360192 | 0.88 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr20_+_50052627 | 0.84 |
ENSDART00000188799
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr7_+_24528430 | 0.83 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr19_-_35439237 | 0.83 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr1_+_53321878 | 0.80 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr7_-_33868903 | 0.78 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr2_-_57469115 | 0.76 |
ENSDART00000192201
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr23_+_27779452 | 0.73 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr7_-_53117131 | 0.73 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr23_+_6544453 | 0.72 |
ENSDART00000005373
|
spo11
|
SPO11 meiotic protein covalently bound to DSB |
| chr13_-_36525982 | 0.70 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr23_+_26026383 | 0.70 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr22_-_38274188 | 0.69 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr7_+_20917966 | 0.69 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr1_-_24349759 | 0.69 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr23_+_27778670 | 0.68 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr5_+_22974019 | 0.68 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr19_+_791538 | 0.67 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr6_+_1724889 | 0.66 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr23_+_2728095 | 0.64 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr5_-_54712159 | 0.63 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr1_+_24469313 | 0.63 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr20_-_43723860 | 0.63 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr5_+_6670945 | 0.62 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr17_+_24590177 | 0.61 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr14_-_26482096 | 0.61 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr3_-_60027255 | 0.60 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr4_-_17263210 | 0.59 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr12_-_7234915 | 0.57 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr23_+_14771979 | 0.57 |
ENSDART00000137410
|
exosc10
|
exosome component 10 |
| chr13_+_2894536 | 0.57 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr7_+_74141297 | 0.56 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr22_-_16758438 | 0.56 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr6_-_55399214 | 0.55 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr5_-_57204352 | 0.55 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr12_-_30359031 | 0.55 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr11_-_11791718 | 0.55 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr4_+_72235562 | 0.53 |
ENSDART00000168547
|
si:cabz01071912.2
|
si:cabz01071912.2 |
| chr18_+_45666489 | 0.53 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr21_+_39197418 | 0.53 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr14_+_30285613 | 0.52 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr21_-_21465111 | 0.51 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr8_-_3312384 | 0.50 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr15_+_20239141 | 0.50 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr12_-_4243268 | 0.50 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr21_-_21515466 | 0.50 |
ENSDART00000147593
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr21_-_21178410 | 0.48 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr2_-_43851915 | 0.47 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr22_+_30195257 | 0.47 |
ENSDART00000027803
ENSDART00000172496 |
add3a
|
adducin 3 (gamma) a |
| chr5_+_18014931 | 0.46 |
ENSDART00000142562
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr11_-_25853212 | 0.46 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr2_-_43852207 | 0.46 |
ENSDART00000192627
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr12_-_30358836 | 0.46 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr5_+_50913357 | 0.45 |
ENSDART00000092938
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr18_-_26781616 | 0.45 |
ENSDART00000136776
ENSDART00000076484 |
kti12
|
KTI12 chromatin associated homolog |
| chr2_-_23677422 | 0.45 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr18_-_38271298 | 0.44 |
ENSDART00000143016
|
caprin1b
|
cell cycle associated protein 1b |
| chr25_+_9027831 | 0.44 |
ENSDART00000155855
|
im:7145024
|
im:7145024 |
| chr23_+_2560005 | 0.44 |
ENSDART00000186906
|
GGT7
|
gamma-glutamyltransferase 7 |
| chr10_+_37173029 | 0.44 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr12_-_2869565 | 0.43 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr17_-_49412313 | 0.43 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr7_+_5975194 | 0.43 |
ENSDART00000123660
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr3_-_15080226 | 0.43 |
ENSDART00000109818
ENSDART00000139835 |
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr20_-_33556983 | 0.43 |
ENSDART00000168798
|
rock2bl
|
rho-associated, coiled-coil containing protein kinase 2b, like |
| chr20_-_3238110 | 0.42 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr2_-_58075414 | 0.42 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr21_-_2958422 | 0.42 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr21_+_39197628 | 0.42 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr9_-_28990649 | 0.42 |
ENSDART00000078823
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr23_+_39963599 | 0.41 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr17_-_11151655 | 0.41 |
ENSDART00000156383
|
CU179699.1
|
|
| chr21_-_2287589 | 0.41 |
ENSDART00000161554
|
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr18_+_13077800 | 0.40 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr5_+_42259002 | 0.40 |
ENSDART00000083778
|
eral1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr5_+_31049742 | 0.40 |
ENSDART00000173097
|
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr2_+_29257942 | 0.40 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr11_-_43473824 | 0.40 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr22_+_9922301 | 0.39 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr8_+_8196087 | 0.39 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr4_+_31456734 | 0.39 |
ENSDART00000140176
|
znf996
|
zinc finger protein 996 |
| chr16_+_26747766 | 0.39 |
ENSDART00000183257
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr3_-_55511569 | 0.39 |
ENSDART00000108995
|
tex2
|
testis expressed 2 |
| chr16_+_27444098 | 0.38 |
ENSDART00000157690
|
invs
|
inversin |
| chr9_+_45428041 | 0.38 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr10_-_33588106 | 0.38 |
ENSDART00000155634
|
eva1c
|
eva-1 homolog C (C. elegans) |
| chr14_+_989733 | 0.38 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr3_+_40255408 | 0.38 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr22_-_26251563 | 0.38 |
ENSDART00000060888
ENSDART00000142821 |
ccdc130
|
coiled-coil domain containing 130 |
| chr2_+_23677179 | 0.38 |
ENSDART00000153918
|
oxsr1a
|
oxidative stress responsive 1a |
| chr2_+_26237322 | 0.38 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr10_-_3416258 | 0.37 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr4_+_77993977 | 0.37 |
ENSDART00000174118
|
terfa
|
telomeric repeat binding factor a |
| chr4_+_26053044 | 0.37 |
ENSDART00000039877
|
SCYL2
|
si:ch211-244b2.1 |
| chr14_-_21618005 | 0.37 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr22_+_2657533 | 0.37 |
ENSDART00000034414
|
zgc:110821
|
zgc:110821 |
| chr3_+_15773991 | 0.36 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr1_+_56180416 | 0.36 |
ENSDART00000089358
|
crb3b
|
crumbs homolog 3b |
| chr3_+_7040363 | 0.36 |
ENSDART00000157805
|
BX000701.2
|
|
| chr5_-_68244564 | 0.35 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr10_-_7472323 | 0.35 |
ENSDART00000163702
ENSDART00000167054 ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr20_+_54336137 | 0.35 |
ENSDART00000113792
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr7_-_22632518 | 0.35 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr5_+_30596822 | 0.34 |
ENSDART00000188375
|
hinfp
|
histone H4 transcription factor |
| chr22_+_7480465 | 0.34 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr7_+_9041174 | 0.34 |
ENSDART00000163411
|
AL935128.1
|
|
| chr11_-_45470370 | 0.34 |
ENSDART00000173098
|
gemin6
|
gem (nuclear organelle) associated protein 6 |
| chr7_-_22632690 | 0.34 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr15_-_23761580 | 0.34 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr4_-_4795205 | 0.32 |
ENSDART00000039313
|
zgc:162331
|
zgc:162331 |
| chr13_-_11800281 | 0.32 |
ENSDART00000079392
|
zmp:0000000662
|
zmp:0000000662 |
| chr5_+_431994 | 0.32 |
ENSDART00000181692
ENSDART00000170350 |
thap1
|
THAP domain containing, apoptosis associated protein 1 |
| chr25_-_22889519 | 0.32 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr3_-_32362872 | 0.32 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr8_-_1698155 | 0.32 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr1_-_59313465 | 0.32 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr4_-_2162688 | 0.32 |
ENSDART00000148900
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr22_-_12304591 | 0.31 |
ENSDART00000136408
|
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr22_+_22438783 | 0.31 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr13_+_35955562 | 0.31 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr20_+_32756763 | 0.31 |
ENSDART00000023006
|
fam84a
|
family with sequence similarity 84, member A |
| chr3_-_42016693 | 0.31 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr10_-_33588310 | 0.31 |
ENSDART00000108658
|
eva1c
|
eva-1 homolog C (C. elegans) |
| chr19_-_307246 | 0.31 |
ENSDART00000145916
|
lingo4a
|
leucine rich repeat and Ig domain containing 4a |
| chr3_-_30888415 | 0.31 |
ENSDART00000124458
|
kmt5c
|
lysine methyltransferase 5C |
| chr18_+_6641542 | 0.31 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr10_-_94184 | 0.31 |
ENSDART00000146125
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr2_-_7605121 | 0.31 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr12_+_30705769 | 0.31 |
ENSDART00000186448
ENSDART00000066259 |
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr2_+_19236969 | 0.30 |
ENSDART00000163875
ENSDART00000168644 |
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr5_-_38384289 | 0.30 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr19_+_4061699 | 0.30 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr20_+_36806398 | 0.30 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr8_-_38022298 | 0.30 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr7_+_14005111 | 0.30 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr17_-_15777350 | 0.30 |
ENSDART00000155711
|
ankrd6a
|
ankyrin repeat domain 6a |
| chr2_+_58877162 | 0.30 |
ENSDART00000122174
|
CABZ01085658.1
|
|
| chr22_-_22231720 | 0.30 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr5_-_12031174 | 0.30 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr13_+_1381953 | 0.29 |
ENSDART00000019983
|
rab23
|
RAB23, member RAS oncogene family |
| chr7_-_19369002 | 0.29 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr21_-_35325466 | 0.29 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr17_-_24575893 | 0.29 |
ENSDART00000141914
|
aftphb
|
aftiphilin b |
| chr11_-_45429199 | 0.29 |
ENSDART00000173111
|
rfc4
|
replication factor C (activator 1) 4 |
| chr21_+_198269 | 0.29 |
ENSDART00000193485
ENSDART00000163696 |
gsdf
|
gonadal somatic cell derived factor |
| chr24_-_31904924 | 0.29 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr10_-_44467384 | 0.29 |
ENSDART00000186382
|
CABZ01072089.1
|
|
| chr3_+_37827373 | 0.29 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr19_+_4068134 | 0.29 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr6_+_7555614 | 0.29 |
ENSDART00000187590
|
myh10
|
myosin, heavy chain 10, non-muscle |
| chr21_-_217589 | 0.28 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr9_+_45493341 | 0.28 |
ENSDART00000145616
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr24_+_36018164 | 0.28 |
ENSDART00000182815
ENSDART00000126941 |
gnal2
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type 2 |
| chr25_+_32390794 | 0.28 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr4_-_74109792 | 0.28 |
ENSDART00000174320
|
RAB21
|
zgc:154045 |
| chr15_+_38221038 | 0.27 |
ENSDART00000188149
|
stim1a
|
stromal interaction molecule 1a |
| chr15_-_434503 | 0.27 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr1_-_7673376 | 0.27 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr4_-_60312069 | 0.27 |
ENSDART00000167207
|
si:dkey-248e17.4
|
si:dkey-248e17.4 |
| chr23_-_27607039 | 0.27 |
ENSDART00000183639
|
phf8
|
PHD finger protein 8 |
| chr4_+_57505842 | 0.27 |
ENSDART00000127643
|
BX510934.1
|
|
| chr1_-_40341306 | 0.27 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr20_+_30797329 | 0.27 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr12_+_30706158 | 0.26 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr1_+_10019653 | 0.26 |
ENSDART00000190227
|
trim2b
|
tripartite motif containing 2b |
| chr3_+_17456428 | 0.25 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr20_-_33583779 | 0.25 |
ENSDART00000097823
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr7_-_8738827 | 0.25 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr16_+_33163858 | 0.25 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr5_-_32505109 | 0.25 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr10_+_5954787 | 0.25 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr15_+_38221196 | 0.25 |
ENSDART00000122134
ENSDART00000190099 |
stim1a
|
stromal interaction molecule 1a |
| chr6_+_12482599 | 0.24 |
ENSDART00000090316
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr13_-_24669258 | 0.24 |
ENSDART00000000831
|
znf511
|
zinc finger protein 511 |
| chr7_+_38090515 | 0.24 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr9_-_28255029 | 0.24 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr6_-_10728057 | 0.23 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr10_-_108952 | 0.23 |
ENSDART00000127228
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr22_+_2417105 | 0.23 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr16_+_41060161 | 0.23 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr24_+_19578935 | 0.23 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr6_+_58289335 | 0.22 |
ENSDART00000177399
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr13_+_7578111 | 0.22 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr15_-_16177603 | 0.22 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of snai2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 4.4 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.9 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 0.8 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 1.7 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.2 | 0.6 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.2 | 0.6 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.2 | 1.4 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.7 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 1.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.4 | GO:0036135 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.7 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.3 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.0 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.4 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.6 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.2 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.3 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.7 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.4 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.6 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0060406 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 1.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.6 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.7 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.7 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 1.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.7 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.7 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 1.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.7 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.9 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |