Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
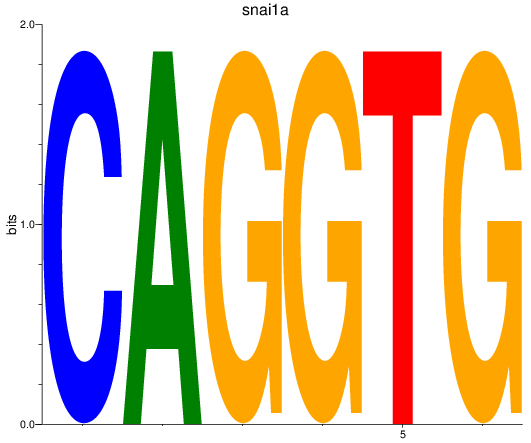
Results for snai1a
Z-value: 3.50
Transcription factors associated with snai1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai1a
|
ENSDARG00000056995 | snail family zinc finger 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai1a | dr11_v1_chr11_-_25257595_25257595 | 0.03 | 9.3e-01 | Click! |
Activity profile of snai1a motif
Sorted Z-values of snai1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_907266 | 5.35 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr23_+_553396 | 4.36 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr7_+_67699178 | 4.13 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr22_+_17828267 | 3.91 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr12_-_4301234 | 3.82 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr5_-_15494164 | 3.81 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr6_+_40523370 | 3.38 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr23_+_2728095 | 3.24 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr7_-_55648336 | 3.24 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr1_+_2712956 | 3.23 |
ENSDART00000126093
|
gpc6a
|
glypican 6a |
| chr11_-_43473824 | 3.21 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr15_+_29025090 | 3.20 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr20_-_43743700 | 3.20 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr17_-_38887424 | 3.16 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr14_+_15155684 | 3.15 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr21_-_34261677 | 3.01 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr7_+_67699009 | 3.00 |
ENSDART00000192810
|
zgc:162592
|
zgc:162592 |
| chr6_+_153146 | 2.98 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr1_+_53321878 | 2.97 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr13_+_31716820 | 2.96 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr5_-_33236637 | 2.94 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr14_+_22457230 | 2.92 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr4_-_2196798 | 2.84 |
ENSDART00000110178
ENSDART00000149330 |
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr7_+_24528430 | 2.81 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr15_-_19128705 | 2.77 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr4_-_17263210 | 2.77 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr14_+_30285613 | 2.76 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr3_+_43086548 | 2.76 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr23_-_44574059 | 2.74 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr23_+_39854566 | 2.72 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr21_-_43666420 | 2.70 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr18_-_50845804 | 2.70 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr9_-_32912638 | 2.68 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr19_+_206835 | 2.65 |
ENSDART00000161137
|
scn1bb
|
sodium channel, voltage-gated, type I, beta b |
| chr15_+_29024895 | 2.64 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr11_-_13152215 | 2.62 |
ENSDART00000160989
ENSDART00000158239 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr22_-_22340688 | 2.60 |
ENSDART00000105597
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr12_-_23365737 | 2.49 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr12_+_13091842 | 2.47 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr6_-_12912606 | 2.46 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr1_-_40341306 | 2.44 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr19_-_18313303 | 2.42 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr8_-_1698155 | 2.40 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr14_-_35672890 | 2.40 |
ENSDART00000074710
|
pdgfc
|
platelet derived growth factor c |
| chr7_+_58751504 | 2.40 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr10_+_40324395 | 2.40 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr20_+_21268795 | 2.39 |
ENSDART00000090016
|
nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr13_-_36525982 | 2.39 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr14_-_46198373 | 2.38 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr17_+_1360192 | 2.34 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr15_-_43164591 | 2.33 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr23_+_43684494 | 2.31 |
ENSDART00000149878
|
otud4
|
OTU deubiquitinase 4 |
| chr2_+_45696743 | 2.28 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr9_+_45428041 | 2.27 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr16_-_25233515 | 2.26 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr8_-_38022298 | 2.26 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr1_+_24387659 | 2.24 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr16_-_42066523 | 2.23 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr3_-_32831429 | 2.22 |
ENSDART00000184932
|
zgc:153733
|
zgc:153733 |
| chr5_-_30080332 | 2.19 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr11_+_18175893 | 2.17 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr14_-_51619388 | 2.17 |
ENSDART00000170949
|
unc5a
|
unc-5 netrin receptor A |
| chr16_-_31598771 | 2.17 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr5_-_11809404 | 2.14 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr15_+_44929497 | 2.12 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr7_-_51773166 | 2.11 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr1_-_34447515 | 2.11 |
ENSDART00000143048
|
lmo7b
|
LIM domain 7b |
| chr16_+_51462620 | 2.11 |
ENSDART00000169443
|
SLC9A1
|
solute carrier family 9 member A1 |
| chr6_+_10333920 | 2.08 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr4_-_4507761 | 2.08 |
ENSDART00000130588
|
tbc1d30
|
TBC1 domain family, member 30 |
| chr14_+_28281744 | 2.07 |
ENSDART00000173292
|
mid2
|
midline 2 |
| chr5_+_57924611 | 2.07 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr17_-_25331439 | 2.06 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr5_+_20255568 | 2.06 |
ENSDART00000153643
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr9_+_33236844 | 2.04 |
ENSDART00000138871
ENSDART00000110066 |
si:dkey-216e24.3
|
si:dkey-216e24.3 |
| chr23_-_28120058 | 2.04 |
ENSDART00000087815
|
b4galnt1a
|
beta-1,4-N-acetyl-galactosaminyl transferase 1a |
| chr23_+_28128453 | 2.03 |
ENSDART00000182618
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr13_-_36911118 | 2.02 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr1_-_49521407 | 1.99 |
ENSDART00000189845
ENSDART00000143474 |
zp3c
|
zona pellucida glycoprotein 3c |
| chr3_-_1317290 | 1.99 |
ENSDART00000047094
|
LO018552.1
|
|
| chr10_+_1052591 | 1.99 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr11_+_41560792 | 1.98 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr3_+_18447574 | 1.95 |
ENSDART00000182342
|
tbc1d16
|
TBC1 domain family, member 16 |
| chr11_-_13151841 | 1.93 |
ENSDART00000161983
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr11_+_18130300 | 1.93 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr22_-_16758438 | 1.92 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr10_+_37173029 | 1.92 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr22_+_26600834 | 1.91 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr10_-_25217347 | 1.90 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr2_+_27855102 | 1.90 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr18_-_25771553 | 1.88 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr19_-_307246 | 1.88 |
ENSDART00000145916
|
lingo4a
|
leucine rich repeat and Ig domain containing 4a |
| chr23_-_32156278 | 1.87 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr15_+_38299385 | 1.87 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr1_+_24469313 | 1.84 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr10_+_31809226 | 1.84 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr15_-_25269028 | 1.83 |
ENSDART00000078230
ENSDART00000193872 |
mettl16
|
methyltransferase like 16 |
| chr18_-_5781922 | 1.83 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr6_+_49028874 | 1.82 |
ENSDART00000175254
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr1_-_6085750 | 1.81 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr3_+_37827373 | 1.80 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr10_+_37181780 | 1.80 |
ENSDART00000187625
|
ksr1a
|
kinase suppressor of ras 1a |
| chr6_+_515181 | 1.79 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr2_+_27855346 | 1.78 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr13_+_7578111 | 1.78 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr12_-_17810543 | 1.77 |
ENSDART00000090484
|
tecpr1a
|
tectonin beta-propeller repeat containing 1a |
| chr6_-_52484566 | 1.77 |
ENSDART00000112146
|
fam83c
|
family with sequence similarity 83, member C |
| chr25_+_9027831 | 1.76 |
ENSDART00000155855
|
im:7145024
|
im:7145024 |
| chr18_-_40708537 | 1.75 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr2_-_7605121 | 1.73 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr21_-_30293224 | 1.71 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr3_-_60027255 | 1.71 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr20_+_25486021 | 1.71 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr14_-_21618005 | 1.71 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr9_-_8296723 | 1.70 |
ENSDART00000139867
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr10_+_19596214 | 1.70 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr9_+_28232522 | 1.70 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr12_-_33359052 | 1.70 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr18_-_26675699 | 1.69 |
ENSDART00000113280
|
FRMD5
|
si:ch211-69m14.1 |
| chr13_+_28618086 | 1.68 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr8_-_17771755 | 1.67 |
ENSDART00000063592
|
prkcz
|
protein kinase C, zeta |
| chr21_-_21465111 | 1.67 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr10_-_31805923 | 1.66 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr25_-_17587785 | 1.66 |
ENSDART00000073679
ENSDART00000146851 |
zgc:66449
|
zgc:66449 |
| chr24_+_12835935 | 1.66 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr3_+_18840810 | 1.66 |
ENSDART00000181137
ENSDART00000128626 ENSDART00000133332 |
tmem104
|
transmembrane protein 104 |
| chr11_-_18800299 | 1.65 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr4_-_17278904 | 1.64 |
ENSDART00000178686
ENSDART00000135730 |
lrmp
|
lymphoid-restricted membrane protein |
| chr10_+_5954787 | 1.63 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr9_+_20780813 | 1.63 |
ENSDART00000142787
|
fam46c
|
family with sequence similarity 46, member C |
| chr9_+_20781047 | 1.62 |
ENSDART00000139174
|
fam46c
|
family with sequence similarity 46, member C |
| chr13_+_40815012 | 1.62 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr17_-_36860988 | 1.62 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr20_+_54336137 | 1.61 |
ENSDART00000113792
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr7_-_36113098 | 1.61 |
ENSDART00000064913
|
fto
|
fat mass and obesity associated |
| chr20_-_26846028 | 1.61 |
ENSDART00000136687
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr18_-_50842155 | 1.61 |
ENSDART00000174139
|
PDPR
|
si:cabz01113374.3 |
| chr17_-_5610514 | 1.61 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr6_+_1724889 | 1.61 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr12_+_20691310 | 1.60 |
ENSDART00000064335
|
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr5_-_64355227 | 1.60 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr13_+_2448251 | 1.60 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr2_+_52232630 | 1.59 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr14_-_7409364 | 1.59 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr3_-_3439150 | 1.59 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr12_-_10512911 | 1.58 |
ENSDART00000124562
ENSDART00000106163 |
zgc:152977
|
zgc:152977 |
| chr3_+_17456428 | 1.58 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr3_+_42923275 | 1.58 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr9_+_46745348 | 1.57 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr7_-_48251234 | 1.57 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr2_-_15040345 | 1.56 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr6_+_58289335 | 1.56 |
ENSDART00000177399
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr2_+_29995590 | 1.56 |
ENSDART00000151906
|
rbm33b
|
RNA binding motif protein 33b |
| chr17_+_24936059 | 1.56 |
ENSDART00000082438
|
dlgap2a
|
discs, large (Drosophila) homolog-associated protein 2a |
| chr3_+_1179601 | 1.55 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr20_-_3997531 | 1.55 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr19_-_977849 | 1.55 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr6_+_45692026 | 1.55 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr12_+_22674030 | 1.55 |
ENSDART00000153254
ENSDART00000152930 |
cdca9
|
cell division cycle associated 9 |
| chr10_-_44411032 | 1.55 |
ENSDART00000111509
|
CABZ01072096.1
|
|
| chr21_-_7940043 | 1.54 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr8_+_45334255 | 1.54 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr22_-_17652112 | 1.54 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr14_-_14659023 | 1.54 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr21_-_21178410 | 1.53 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr18_+_8917766 | 1.53 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr5_+_44846434 | 1.53 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr12_-_18483348 | 1.53 |
ENSDART00000152757
|
tex2l
|
testis expressed 2, like |
| chr15_+_38299563 | 1.52 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr24_-_24271629 | 1.52 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr7_-_48263516 | 1.51 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr13_+_35955562 | 1.51 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr8_-_21184759 | 1.51 |
ENSDART00000139257
|
gls2a
|
glutaminase 2a (liver, mitochondrial) |
| chr1_-_24349759 | 1.51 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr5_+_22974019 | 1.51 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr5_-_23715861 | 1.50 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr7_-_19369002 | 1.50 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr11_+_6882362 | 1.49 |
ENSDART00000144181
|
klhl26
|
kelch-like family member 26 |
| chr7_+_39679944 | 1.49 |
ENSDART00000133420
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr1_+_51391082 | 1.49 |
ENSDART00000063936
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr6_-_40449399 | 1.49 |
ENSDART00000103879
|
tatdn2
|
TatD DNase domain containing 2 |
| chr20_-_14114078 | 1.47 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr15_-_33964897 | 1.47 |
ENSDART00000172075
ENSDART00000158126 ENSDART00000160456 |
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr2_+_30531726 | 1.46 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr23_-_26488622 | 1.46 |
ENSDART00000168052
ENSDART00000169088 |
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr21_-_21515466 | 1.46 |
ENSDART00000147593
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr10_-_7734939 | 1.46 |
ENSDART00000075563
|
snx24
|
sorting nexin 24 |
| chr3_+_32416948 | 1.45 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr2_-_50225411 | 1.45 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr5_-_13848296 | 1.45 |
ENSDART00000127109
|
figla
|
folliculogenesis specific bHLH transcription factor |
| chr12_+_46543572 | 1.45 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr7_+_69449814 | 1.45 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr7_-_33868903 | 1.45 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr16_+_39159752 | 1.45 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr11_-_669558 | 1.44 |
ENSDART00000173450
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr19_-_35439237 | 1.44 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr11_+_18037729 | 1.44 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr21_+_26748141 | 1.44 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr5_-_31773208 | 1.44 |
ENSDART00000137556
ENSDART00000122066 |
fam102ab
|
family with sequence similarity 102, member Ab |
| chr17_+_23549117 | 1.44 |
ENSDART00000063953
|
zgc:65997
|
zgc:65997 |
Network of associatons between targets according to the STRING database.
First level regulatory network of snai1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0035046 | pronuclear migration(GO:0035046) |
| 1.3 | 3.9 | GO:0030237 | female sex determination(GO:0030237) |
| 0.9 | 3.6 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.8 | 1.6 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.8 | 13.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.7 | 2.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.7 | 5.5 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.7 | 4.7 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.7 | 2.6 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.6 | 3.7 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.6 | 1.8 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.5 | 2.7 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.5 | 2.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.5 | 1.6 | GO:0042245 | RNA repair(GO:0042245) |
| 0.5 | 2.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.5 | 2.6 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.5 | 2.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.5 | 2.5 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.5 | 2.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.5 | 1.5 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.5 | 1.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.5 | 2.9 | GO:0044819 | mitotic G1/S transition checkpoint(GO:0044819) |
| 0.5 | 1.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.5 | 2.3 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.4 | 1.8 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.4 | 4.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.4 | 1.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.4 | 1.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 1.6 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.4 | 1.6 | GO:0010259 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.4 | 1.6 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 3.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.4 | 1.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.4 | 0.8 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.4 | 2.2 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.4 | 2.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.4 | 1.1 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.4 | 2.5 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.4 | 1.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.4 | 1.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 1.4 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.4 | 3.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.4 | 4.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.3 | 3.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 2.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 3.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 1.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 1.3 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.3 | 1.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.3 | 1.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 1.3 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.3 | 2.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.3 | 1.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 1.6 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.3 | 1.3 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.3 | 2.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 6.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.3 | 1.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.3 | 2.4 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.3 | 0.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.3 | 2.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.3 | 0.9 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 3.5 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.3 | 1.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.3 | 1.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.3 | 1.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 2.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.3 | 2.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 0.9 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.3 | 0.8 | GO:0015824 | proline transport(GO:0015824) |
| 0.3 | 1.9 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.3 | 2.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.3 | 0.8 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.3 | 4.9 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.3 | 5.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 1.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.3 | 0.8 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.3 | 1.0 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.3 | 0.8 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.2 | 0.7 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.2 | 3.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.7 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.2 | 1.2 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.2 | 1.7 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.2 | 1.0 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 0.7 | GO:0036135 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.2 | 1.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.7 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 3.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 1.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 0.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 2.1 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.2 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 2.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.7 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 3.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 0.9 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 0.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 2.7 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 0.9 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.2 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 1.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 2.6 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.2 | 2.0 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 3.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.6 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.2 | 1.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 1.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 1.0 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 2.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 2.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 0.2 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.2 | 1.0 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 1.0 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 0.8 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 0.6 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 1.0 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 4.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.8 | GO:0035521 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 1.0 | GO:0010990 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 0.6 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.2 | 1.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 1.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 1.7 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.2 | 0.8 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 2.1 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.2 | 2.4 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.2 | 0.4 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.2 | 3.0 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.2 | 2.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 2.9 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 1.4 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.2 | 1.4 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.7 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 1.6 | GO:0008585 | female gonad development(GO:0008585) |
| 0.2 | 1.7 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.2 | 1.4 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.2 | 0.3 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 2.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 1.0 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.2 | 0.3 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 5.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 3.9 | GO:0048599 | oocyte development(GO:0048599) |
| 0.2 | 1.6 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 1.6 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 1.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 2.8 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.2 | 1.0 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 0.6 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 0.6 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.2 | 6.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.2 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 1.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 1.2 | GO:1900026 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 19.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.2 | 0.8 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.4 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.1 | 2.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 8.6 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 2.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.7 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.6 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 1.8 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 3.8 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.4 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 2.0 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 3.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.7 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.4 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.1 | 2.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.4 | GO:1904375 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.1 | 1.9 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.3 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.1 | 2.5 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 0.8 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 0.8 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 2.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.4 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.5 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.5 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.5 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 1.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.6 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 1.2 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 5.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.7 | GO:1900027 | regulation of ruffle assembly(GO:1900027) positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.8 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.4 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 5.9 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 1.0 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.8 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.1 | 3.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.0 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.5 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 1.0 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.4 | GO:0010668 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.1 | 1.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 2.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.4 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.1 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.5 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.5 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 0.5 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 1.3 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.5 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.5 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 4.3 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.1 | 0.7 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.1 | 2.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 6.4 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 3.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 1.3 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 2.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.9 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.3 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.3 | GO:0045217 | cell-cell junction maintenance(GO:0045217) zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.9 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 1.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.9 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 2.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 3.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.3 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0071314 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.1 | 1.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.6 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 2.6 | GO:0043266 | regulation of potassium ion transport(GO:0043266) |
| 0.1 | 0.2 | GO:1902176 | negative regulation of cellular response to oxidative stress(GO:1900408) regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) negative regulation of response to oxidative stress(GO:1902883) regulation of oxidative stress-induced cell death(GO:1903201) negative regulation of oxidative stress-induced cell death(GO:1903202) |
| 0.1 | 0.6 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0071331 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 1.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.4 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.3 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.1 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 5.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 0.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.6 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 8.4 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.1 | 0.7 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 0.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.0 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 1.4 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 2.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 2.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 2.4 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.1 | 1.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.8 | GO:0045161 | neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 7.1 | GO:0051494 | negative regulation of cytoskeleton organization(GO:0051494) |
| 0.1 | 0.3 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 3.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.2 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 1.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.4 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.5 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 12.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 0.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.2 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.5 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 3.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.2 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.1 | 3.2 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.1 | 0.5 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.1 | 0.2 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 1.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.3 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.1 | 0.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.5 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.4 | GO:0032366 | intracellular lipid transport(GO:0032365) intracellular sterol transport(GO:0032366) |
| 0.1 | 0.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.6 | GO:0048885 | neuromast deposition(GO:0048885) |
| 0.1 | 0.7 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.7 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 3.4 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.5 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 1.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.5 | GO:0090311 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.1 | 5.1 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.1 | 0.2 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 0.2 | GO:0042308 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.4 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.2 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 0.9 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 0.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.5 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 1.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 1.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.3 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 1.3 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.2 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.2 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 1.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 3.0 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.8 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.6 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0042726 | FAD biosynthetic process(GO:0006747) flavin-containing compound metabolic process(GO:0042726) flavin-containing compound biosynthetic process(GO:0042727) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.2 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.0 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.6 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.4 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.8 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 2.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.0 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 | 2.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.5 | GO:2001242 | regulation of intrinsic apoptotic signaling pathway(GO:2001242) negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.4 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.0 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 1.3 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 2.0 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) thymocyte migration(GO:0072679) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 1.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.5 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.5 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.5 | GO:0048016 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 1.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.1 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 2.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 1.7 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.4 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.5 | GO:0071902 | positive regulation of protein serine/threonine kinase activity(GO:0071902) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.0 | 6.3 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 1.0 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 4.8 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 2.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.3 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 2.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.7 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0060471 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 1.9 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 1.1 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 1.5 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 1.3 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.9 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.5 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 3.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0044774 | mitotic DNA integrity checkpoint(GO:0044774) |
| 0.0 | 1.6 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 1.3 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.7 | GO:0006304 | DNA modification(GO:0006304) |
| 0.0 | 0.5 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 2.0 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.5 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 2.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.7 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.9 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.7 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.3 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.2 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.1 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0043331 | response to dsRNA(GO:0043331) |
| 0.0 | 0.5 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 1.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.9 | 2.6 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.6 | 3.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 1.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.5 | 1.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.5 | 2.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.5 | 2.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.5 | 2.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.5 | 3.7 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.4 | 1.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 1.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.4 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.4 | 4.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.4 | 6.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 2.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 1.4 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.3 | 1.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.3 | 4.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.3 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 1.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 2.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 0.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 0.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 1.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.4 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 1.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 3.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 2.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 1.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 0.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 1.9 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 3.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 6.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.2 | 1.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.8 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.2 | 1.8 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.2 | 0.5 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.2 | 3.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 3.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.7 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 2.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 2.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.6 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 3.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.5 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 1.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.3 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 15.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 5.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 1.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 7.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.4 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 2.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.6 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 3.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) ribonuclease P complex(GO:0030677) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 7.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 5.5 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 4.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 7.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.6 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 3.9 | GO:0044440 | endosomal part(GO:0044440) |
| 0.1 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.8 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 3.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 12.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 6.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 3.0 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 2.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 10.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 5.8 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 3.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 17.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.1 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.5 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.9 | 7.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.9 | 2.6 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.8 | 13.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.8 | 3.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.7 | 5.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.7 | 2.6 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.6 | 1.8 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.6 | 2.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.5 | 2.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.5 | 2.0 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.5 | 2.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.5 | 4.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.5 | 1.5 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.5 | 1.9 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.5 | 2.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.5 | 2.3 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.4 | 1.2 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.4 | 2.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.4 | 3.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.4 | 1.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.4 | 11.0 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.4 | 1.6 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.4 | 4.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.4 | 1.1 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 1.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.4 | 2.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 1.4 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.3 | 3.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 3.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 1.3 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.3 | 1.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 1.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 2.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.3 | 1.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.3 | 2.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 2.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 0.9 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 1.5 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 0.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 2.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 0.9 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.3 | 3.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 3.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 2.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 2.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 5.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 1.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.3 | 4.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 3.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 1.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 3.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 0.7 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 1.0 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 3.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 2.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 2.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 0.7 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.2 | 1.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 0.7 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.2 | 4.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 2.0 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 1.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.9 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.2 | 1.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 2.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 0.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 1.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 2.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 2.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 2.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 0.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.6 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.2 | 1.6 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 1.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 5.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.7 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 3.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 0.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 1.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 4.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 2.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 5.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 0.9 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 1.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 1.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 0.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 0.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 0.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 0.6 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.2 | 1.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 2.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 2.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.0 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.0 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 2.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 3.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 1.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 1.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 2.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.4 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 2.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 1.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 3.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 1.4 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.1 | 4.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.7 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 1.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 1.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.6 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.6 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.7 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 2.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.6 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.4 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 1.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.9 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.7 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 2.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 1.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.9 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 4.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 1.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 2.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 2.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 3.2 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 0.4 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.9 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 2.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.3 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.1 | 1.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 1.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.5 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 3.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 3.9 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 0.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.6 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 2.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 3.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 27.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 5.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 7.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 2.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.4 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.5 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 1.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.5 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 3.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.0 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 2.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 4.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 2.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 1.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 4.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 3.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters(GO:0016894) |
| 0.0 | 1.1 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 10.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.8 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 3.6 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.5 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 2.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.6 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 1.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 3.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 2.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 3.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.3 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 3.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 10.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.6 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 2.4 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.1 | GO:0004777 | succinate-semialdehyde dehydrogenase (NAD+) activity(GO:0004777) succinate-semialdehyde dehydrogenase [NAD(P)+] activity(GO:0009013) |
| 0.0 | 0.1 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 5.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 1.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.9 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.2 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 3.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 12.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.9 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 13.2 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 16.6 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 1.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.7 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0071916 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 4.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 6.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 2.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 3.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 2.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 4.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 1.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 4.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 2.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 4.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 4.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 2.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 1.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 2.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 1.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 1.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.2 | 1.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.6 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 4.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 3.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 2.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.9 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.1 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.1 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.8 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 1.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 4.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.2 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 1.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 0.5 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.3 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |