Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
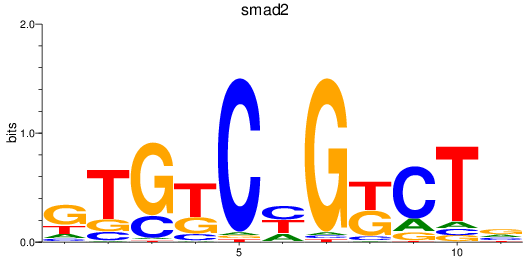
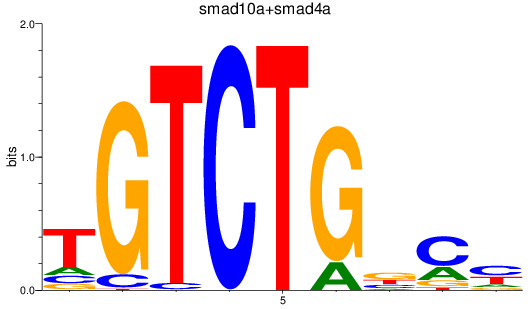
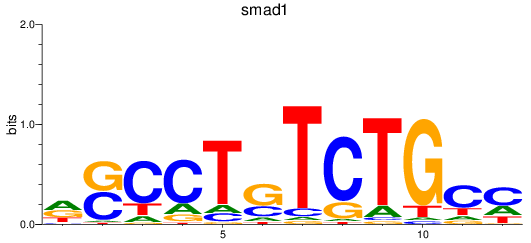
Results for smad2_smad10a+smad4a_smad1
Z-value: 0.42
Transcription factors associated with smad2_smad10a+smad4a_smad1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smad2
|
ENSDARG00000006389 | SMAD family member 2 |
|
smad10a
|
ENSDARG00000045094 | si_dkey-222b8.1 |
|
smad10a
|
ENSDARG00000070428 | si_dkey-222b8.1 |
|
smad4a
|
ENSDARG00000075226 | SMAD family member 4a |
|
smad1
|
ENSDARG00000027199 | SMAD family member 1 |
|
smad1
|
ENSDARG00000112617 | SMAD family member 1 |
|
smad1
|
ENSDARG00000115674 | SMAD family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-239n17.4 | dr11_v1_chr16_-_28878080_28878080 | 0.57 | 1.1e-01 | Click! |
| smad2 | dr11_v1_chr10_-_14920989_14920989 | -0.54 | 1.3e-01 | Click! |
| smad4a | dr11_v1_chr10_+_575929_575929 | -0.47 | 2.0e-01 | Click! |
| smad1 | dr11_v1_chr1_-_35929143_35929143 | 0.45 | 2.3e-01 | Click! |
| si:dkey-222b8.1 | dr11_v1_chr19_+_23932259_23932259 | 0.44 | 2.4e-01 | Click! |
Activity profile of smad2_smad10a+smad4a_smad1 motif
Sorted Z-values of smad2_smad10a+smad4a_smad1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_46040618 | 1.39 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr13_+_25433774 | 0.61 |
ENSDART00000141255
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr9_-_8670158 | 0.42 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr3_-_3496738 | 0.41 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr15_+_40008370 | 0.35 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr19_-_41069573 | 0.31 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr9_+_52411530 | 0.31 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr13_-_5079511 | 0.29 |
ENSDART00000110248
|
slc29a3
|
solute carrier family 29 (equilibrative nucleoside transporter), member 3 |
| chr4_-_16412084 | 0.24 |
ENSDART00000188460
|
dcn
|
decorin |
| chr11_-_497854 | 0.24 |
ENSDART00000104520
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr4_-_4932619 | 0.24 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr14_+_8127893 | 0.22 |
ENSDART00000169091
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr23_-_39849155 | 0.21 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr18_-_44908479 | 0.20 |
ENSDART00000169636
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr3_+_56645710 | 0.19 |
ENSDART00000193978
|
CR759836.1
|
|
| chr11_-_2700397 | 0.19 |
ENSDART00000082511
|
si:ch211-160o17.6
|
si:ch211-160o17.6 |
| chr19_+_42955280 | 0.18 |
ENSDART00000141225
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr20_+_18163355 | 0.18 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr10_+_25272355 | 0.17 |
ENSDART00000140186
|
rab38b
|
RAB38b, member of RAS oncogene family |
| chr24_-_28243186 | 0.17 |
ENSDART00000105691
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr5_-_66028714 | 0.16 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr18_+_49225552 | 0.15 |
ENSDART00000135026
|
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr12_-_4070058 | 0.15 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr19_-_5380770 | 0.14 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr12_-_9294819 | 0.14 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr16_+_31804590 | 0.14 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr17_+_33433576 | 0.13 |
ENSDART00000077581
|
snap23.2
|
synaptosomal-associated protein 23.2 |
| chr8_+_32719930 | 0.13 |
ENSDART00000145362
|
hmcn2
|
hemicentin 2 |
| chr2_+_47623202 | 0.13 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr21_-_45073 | 0.12 |
ENSDART00000185997
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr25_-_31433512 | 0.12 |
ENSDART00000067028
|
zgc:172145
|
zgc:172145 |
| chr12_-_17712393 | 0.11 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr19_+_42219165 | 0.10 |
ENSDART00000163192
|
CU896644.1
|
|
| chr20_-_26042070 | 0.10 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr21_+_26390549 | 0.10 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr25_-_31396479 | 0.10 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr24_-_7699356 | 0.09 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr23_-_44965582 | 0.09 |
ENSDART00000163367
|
tfr2
|
transferrin receptor 2 |
| chr14_-_25949951 | 0.09 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr22_-_11136625 | 0.09 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr23_-_45405968 | 0.09 |
ENSDART00000149462
|
zgc:101853
|
zgc:101853 |
| chr8_+_28527776 | 0.09 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| chr14_+_51098036 | 0.08 |
ENSDART00000184118
|
CABZ01078594.1
|
|
| chr3_+_32526263 | 0.08 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr22_+_635813 | 0.08 |
ENSDART00000179067
|
CU856139.1
|
|
| chr22_+_38824012 | 0.08 |
ENSDART00000144318
ENSDART00000003736 |
anos1b
|
anosmin 1b |
| chr18_+_7345417 | 0.08 |
ENSDART00000041429
|
glipr1b
|
GLI pathogenesis-related 1b |
| chr13_+_281214 | 0.08 |
ENSDART00000137572
|
mpc1
|
mitochondrial pyruvate carrier 1 |
| chr4_-_191736 | 0.08 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr18_-_38087875 | 0.08 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr23_-_24263474 | 0.07 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr23_+_40908583 | 0.07 |
ENSDART00000180933
|
LO017845.1
|
|
| chr14_+_8442248 | 0.07 |
ENSDART00000163059
|
si:dkey-160o24.3
|
si:dkey-160o24.3 |
| chr25_-_210730 | 0.07 |
ENSDART00000187580
|
FP236318.1
|
|
| chr8_-_32497815 | 0.07 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr14_+_6546516 | 0.07 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr7_+_22767678 | 0.07 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr12_+_30653047 | 0.07 |
ENSDART00000148562
|
thbs2b
|
thrombospondin 2b |
| chr5_-_72125551 | 0.06 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr18_+_507618 | 0.06 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr3_-_36846496 | 0.06 |
ENSDART00000055237
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr7_-_72269049 | 0.06 |
ENSDART00000161497
|
CU463038.2
|
|
| chr16_+_40301056 | 0.06 |
ENSDART00000058578
|
rspo3
|
R-spondin 3 |
| chr8_+_47633438 | 0.06 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr24_+_38306010 | 0.06 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr19_-_32710922 | 0.06 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr22_+_30446987 | 0.06 |
ENSDART00000146471
|
si:dkey-169i5.4
|
si:dkey-169i5.4 |
| chr4_+_77740228 | 0.06 |
ENSDART00000193397
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr17_-_122680 | 0.06 |
ENSDART00000066430
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr19_-_5369486 | 0.06 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr4_+_22680442 | 0.06 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr6_-_15653494 | 0.06 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr3_+_14388010 | 0.06 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr20_+_54356540 | 0.06 |
ENSDART00000143591
|
znf410
|
zinc finger protein 410 |
| chr14_-_451555 | 0.06 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr3_+_41714966 | 0.06 |
ENSDART00000155440
|
eif3ba
|
eukaryotic translation initiation factor 3, subunit Ba |
| chr20_-_36809059 | 0.06 |
ENSDART00000062925
|
slc25a27
|
solute carrier family 25, member 27 |
| chr11_+_77526 | 0.06 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr8_+_26292560 | 0.06 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr2_-_3158919 | 0.06 |
ENSDART00000098394
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr19_+_31061718 | 0.06 |
ENSDART00000145971
|
sostdc1b
|
sclerostin domain containing 1b |
| chr12_+_15165736 | 0.05 |
ENSDART00000180398
|
adprh
|
ADP-ribosylarginine hydrolase |
| chr10_+_37137464 | 0.05 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr19_-_36675023 | 0.05 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr5_-_34997630 | 0.05 |
ENSDART00000170684
|
btf3
|
basic transcription factor 3 |
| chr23_+_36106790 | 0.05 |
ENSDART00000128533
|
hoxc3a
|
homeobox C3a |
| chr21_-_7265219 | 0.05 |
ENSDART00000158852
|
egfl7
|
EGF-like-domain, multiple 7 |
| chr22_+_26400519 | 0.05 |
ENSDART00000159839
ENSDART00000144585 |
capn8
|
calpain 8 |
| chr8_-_17516448 | 0.05 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr8_-_46509194 | 0.05 |
ENSDART00000038924
|
sult1st1
|
sulfotransferase family 1, cytosolic sulfotransferase 1 |
| chr8_-_32497581 | 0.05 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr22_-_11137268 | 0.05 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr19_+_25478203 | 0.05 |
ENSDART00000132934
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr9_-_17417628 | 0.05 |
ENSDART00000060425
ENSDART00000141997 |
ftr53
|
finTRIM family, member 53 |
| chr10_+_44042033 | 0.05 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr3_+_32526799 | 0.05 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr1_-_21344478 | 0.05 |
ENSDART00000077805
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr9_-_22339582 | 0.05 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr15_+_33989181 | 0.05 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr4_-_26322834 | 0.05 |
ENSDART00000170709
ENSDART00000161687 |
st8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr4_-_1015896 | 0.05 |
ENSDART00000170292
|
FAM180A
|
family with sequence similarity 180 member A |
| chr14_+_9421510 | 0.05 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr11_+_40032790 | 0.05 |
ENSDART00000158809
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr9_+_23900703 | 0.05 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr1_+_59073203 | 0.05 |
ENSDART00000149937
ENSDART00000162201 |
MFAP4 (1 of many)
zgc:173915
|
si:zfos-2330d3.3 zgc:173915 |
| chr20_+_30445971 | 0.05 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr17_+_22102791 | 0.05 |
ENSDART00000047772
|
mal
|
mal, T cell differentiation protein |
| chr23_-_17470146 | 0.05 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr4_-_18595525 | 0.05 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr20_-_31427390 | 0.05 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr13_+_23157053 | 0.05 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr8_-_2230128 | 0.05 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr20_-_40451115 | 0.05 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr19_-_12322356 | 0.05 |
ENSDART00000016128
|
ncaldb
|
neurocalcin delta b |
| chr1_+_47585700 | 0.05 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr19_-_43819582 | 0.05 |
ENSDART00000160879
ENSDART00000075902 |
klhl43
|
kelch-like family member 43 |
| chr14_-_3381303 | 0.05 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr15_+_9117330 | 0.05 |
ENSDART00000156306
|
ptgir
|
prostaglandin I2 receptor |
| chr14_+_11458044 | 0.05 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr5_-_38451082 | 0.05 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr19_-_47456787 | 0.05 |
ENSDART00000168792
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr20_+_34400715 | 0.05 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr6_-_24103666 | 0.05 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr4_-_9592402 | 0.05 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr2_+_33457310 | 0.05 |
ENSDART00000056657
|
zgc:113531
|
zgc:113531 |
| chr6_+_6491013 | 0.05 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr20_-_7648325 | 0.05 |
ENSDART00000186541
|
CR848049.1
|
|
| chr21_+_30351256 | 0.05 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr25_+_31958911 | 0.05 |
ENSDART00000191394
ENSDART00000090727 ENSDART00000185893 |
duox
|
dual oxidase |
| chr7_-_69025306 | 0.05 |
ENSDART00000180796
|
CABZ01057488.2
|
|
| chr22_+_15507218 | 0.05 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr19_+_19759577 | 0.04 |
ENSDART00000169480
|
hoxa5a
|
homeobox A5a |
| chr2_+_42236118 | 0.04 |
ENSDART00000114162
ENSDART00000141199 |
ftr02
|
finTRIM family, member 2 |
| chr20_+_34326874 | 0.04 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr25_+_31238606 | 0.04 |
ENSDART00000149439
|
tnni2a.2
|
troponin I type 2a (skeletal, fast), tandem duplicate 2 |
| chr16_-_13595027 | 0.04 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr24_-_24282427 | 0.04 |
ENSDART00000123299
|
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr14_-_1949277 | 0.04 |
ENSDART00000159435
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr16_+_5202042 | 0.04 |
ENSDART00000145368
|
soga3a
|
SOGA family member 3a |
| chr2_-_23172708 | 0.04 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr3_-_53559581 | 0.04 |
ENSDART00000183499
|
notch3
|
notch 3 |
| chr22_-_30935510 | 0.04 |
ENSDART00000133335
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr5_+_34997763 | 0.04 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr11_-_42554290 | 0.04 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr15_-_40157165 | 0.04 |
ENSDART00000192991
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr15_-_40157331 | 0.04 |
ENSDART00000187958
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr14_-_12307522 | 0.04 |
ENSDART00000163900
|
myot
|
myotilin |
| chr9_+_21402863 | 0.04 |
ENSDART00000125357
|
cx30.3
|
connexin 30.3 |
| chr25_-_774350 | 0.04 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr3_-_21242460 | 0.04 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr21_-_26495700 | 0.04 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr22_+_17237219 | 0.04 |
ENSDART00000090069
|
axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr3_-_41791178 | 0.04 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr4_+_18963822 | 0.04 |
ENSDART00000066975
ENSDART00000066973 |
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr12_-_43685802 | 0.04 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr18_-_44129151 | 0.04 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| chr4_-_76488854 | 0.04 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr7_-_69857692 | 0.04 |
ENSDART00000124764
|
myoz2a
|
myozenin 2a |
| chr3_-_32320537 | 0.04 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr3_-_46811611 | 0.04 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr3_+_24197934 | 0.04 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr1_-_50710468 | 0.04 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr21_+_28496194 | 0.04 |
ENSDART00000138236
|
cox8a
|
cytochrome c oxidase subunit VIIIA (ubiquitous) |
| chr7_+_20505311 | 0.04 |
ENSDART00000187335
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr17_-_15546862 | 0.04 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr7_+_54642005 | 0.04 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr17_-_36929332 | 0.04 |
ENSDART00000183454
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr17_+_12408188 | 0.04 |
ENSDART00000105218
|
khk
|
ketohexokinase |
| chr16_+_23984179 | 0.04 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr11_+_36180349 | 0.04 |
ENSDART00000012940
|
grm2b
|
glutamate receptor, metabotropic 2b |
| chr4_-_25858620 | 0.04 |
ENSDART00000128968
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr16_+_10918252 | 0.04 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr8_+_16004551 | 0.04 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_-_716426 | 0.04 |
ENSDART00000028159
|
foxf2a
|
forkhead box F2a |
| chr2_-_59265521 | 0.04 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr25_+_14017609 | 0.04 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr19_-_38611814 | 0.04 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr13_+_28854438 | 0.04 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr3_+_26019426 | 0.04 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr14_-_1990290 | 0.04 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr16_+_23961276 | 0.04 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr12_+_47122104 | 0.04 |
ENSDART00000184248
|
CABZ01088982.1
|
|
| chr9_+_310331 | 0.04 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr14_-_40389699 | 0.04 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr11_+_36989696 | 0.04 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr16_+_221739 | 0.04 |
ENSDART00000180243
|
nrsn1
|
neurensin 1 |
| chr19_-_325584 | 0.04 |
ENSDART00000134266
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr10_+_16756546 | 0.04 |
ENSDART00000168624
|
CABZ01024659.1
|
|
| chr4_+_70351760 | 0.04 |
ENSDART00000159832
|
si:dkey-190j3.6
|
si:dkey-190j3.6 |
| chr15_-_40157513 | 0.04 |
ENSDART00000184014
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr13_+_42124566 | 0.03 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr16_-_39859119 | 0.03 |
ENSDART00000138388
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr3_-_53559408 | 0.03 |
ENSDART00000073930
|
notch3
|
notch 3 |
| chr12_-_31103906 | 0.03 |
ENSDART00000189099
ENSDART00000121527 ENSDART00000185635 ENSDART00000183754 |
tcf7l2
|
transcription factor 7 like 2 |
| chr21_+_5993188 | 0.03 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr2_+_26179096 | 0.03 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr18_-_38088099 | 0.03 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr3_+_46635527 | 0.03 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr13_+_23176330 | 0.03 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr5_+_27898226 | 0.03 |
ENSDART00000098604
ENSDART00000180251 |
adam28
|
ADAM metallopeptidase domain 28 |
Network of associatons between targets according to the STRING database.
First level regulatory network of smad2_smad10a+smad4a_smad1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.3 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.0 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |