Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
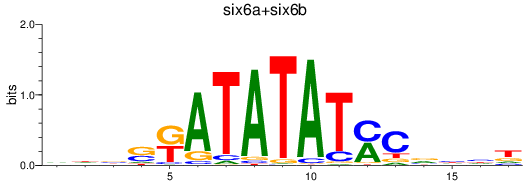
Results for six6a+six6b
Z-value: 0.81
Transcription factors associated with six6a+six6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six6a
|
ENSDARG00000025187 | SIX homeobox 6a |
|
six6b
|
ENSDARG00000031316 | SIX homeobox 6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six6a | dr11_v1_chr13_+_31603988_31603988 | 0.85 | 3.9e-03 | Click! |
| six6b | dr11_v1_chr20_-_20533865_20533865 | 0.84 | 5.0e-03 | Click! |
Activity profile of six6a+six6b motif
Sorted Z-values of six6a+six6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_51098036 | 3.07 |
ENSDART00000184118
|
CABZ01078594.1
|
|
| chr25_+_31277415 | 2.00 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr9_-_22310919 | 1.93 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr13_+_35745572 | 1.88 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr22_-_10459880 | 1.56 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr23_-_42232124 | 1.37 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr2_-_44946094 | 1.26 |
ENSDART00000036997
|
camk2n1a
|
calcium/calmodulin-dependent protein kinase II inhibitor 1a |
| chr14_+_50770537 | 1.19 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr10_-_24648228 | 1.12 |
ENSDART00000081834
ENSDART00000132830 |
stoml3b
|
stomatin (EPB72)-like 3b |
| chr17_+_51627209 | 1.06 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr15_-_1822548 | 0.96 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr20_+_37383034 | 0.96 |
ENSDART00000134997
ENSDART00000145927 |
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr1_+_1789357 | 0.94 |
ENSDART00000006449
|
atp1a1a.2
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 2 |
| chr18_-_14677936 | 0.88 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr2_+_47581997 | 0.87 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr22_+_13977772 | 0.84 |
ENSDART00000080313
|
arl4ca
|
ADP-ribosylation factor-like 4Ca |
| chr16_+_46492756 | 0.83 |
ENSDART00000105931
|
rpz5
|
rapunzel 5 |
| chr13_+_28819768 | 0.82 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr12_+_2446837 | 0.80 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr13_-_16257848 | 0.73 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr3_+_23726148 | 0.70 |
ENSDART00000174580
|
hoxb3a
|
homeobox B3a |
| chr19_-_32710922 | 0.67 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr7_+_20535869 | 0.67 |
ENSDART00000078181
|
zgc:158423
|
zgc:158423 |
| chr13_+_49175624 | 0.65 |
ENSDART00000193550
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr3_+_23731109 | 0.65 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr19_-_17774875 | 0.62 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr21_-_42007213 | 0.61 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr3_+_25154078 | 0.61 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr20_-_9580446 | 0.59 |
ENSDART00000014168
|
zfp36l1b
|
zinc finger protein 36, C3H type-like 1b |
| chr11_-_31039533 | 0.59 |
ENSDART00000127355
|
ier2b
|
immediate early response 2b |
| chr7_+_22767678 | 0.58 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr21_+_51521 | 0.58 |
ENSDART00000007672
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr9_+_907459 | 0.56 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr15_-_42736433 | 0.55 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr16_+_6632969 | 0.55 |
ENSDART00000109276
|
sall3a
|
spalt-like transcription factor 3a |
| chr16_+_46492994 | 0.54 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr13_+_42544009 | 0.54 |
ENSDART00000145409
|
si:dkey-221j11.3
|
si:dkey-221j11.3 |
| chr21_-_25756119 | 0.53 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr11_+_40812590 | 0.53 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr2_-_30249746 | 0.52 |
ENSDART00000124153
ENSDART00000142575 |
elocb
|
elongin C paralog b |
| chr4_+_76637548 | 0.52 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr10_-_25816558 | 0.51 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr6_+_28124393 | 0.51 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr23_-_44226556 | 0.48 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr7_-_31932723 | 0.45 |
ENSDART00000014843
|
bdnf
|
brain-derived neurotrophic factor |
| chr20_+_51104367 | 0.44 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr16_+_9609721 | 0.43 |
ENSDART00000047920
|
enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr22_-_3261879 | 0.43 |
ENSDART00000159643
|
gpr35.1
|
G protein-coupled receptor 35, tandem duplicate 1 |
| chr3_+_50069204 | 0.43 |
ENSDART00000184243
|
zgc:103625
|
zgc:103625 |
| chr10_-_43540196 | 0.42 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr16_+_41570653 | 0.42 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr2_-_27775236 | 0.40 |
ENSDART00000187983
|
XKR4
|
zgc:123035 |
| chr8_-_50259448 | 0.40 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr21_-_3796461 | 0.40 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr15_-_31514818 | 0.40 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr15_-_20939579 | 0.40 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr8_+_52701811 | 0.39 |
ENSDART00000179692
|
bcl2l16
|
BCL2 like 16 |
| chr25_+_3306620 | 0.37 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr18_+_12058403 | 0.37 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr23_+_32335871 | 0.37 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr5_+_64368770 | 0.37 |
ENSDART00000162246
|
plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr19_-_3925801 | 0.36 |
ENSDART00000129570
ENSDART00000163138 |
si:ch73-281i18.6
|
si:ch73-281i18.6 |
| chr21_-_42055872 | 0.36 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr22_+_19640309 | 0.36 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr20_-_49729446 | 0.35 |
ENSDART00000111089
|
filip1b
|
filamin A interacting protein 1b |
| chr19_-_3906369 | 0.35 |
ENSDART00000160299
ENSDART00000169205 |
si:ch73-281i18.4
|
si:ch73-281i18.4 |
| chr14_+_45883687 | 0.33 |
ENSDART00000114790
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr3_+_37707432 | 0.33 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr2_-_30250311 | 0.32 |
ENSDART00000184144
|
elocb
|
elongin C paralog b |
| chr10_+_41549819 | 0.31 |
ENSDART00000114210
|
CABZ01049847.1
|
|
| chr24_-_20016817 | 0.31 |
ENSDART00000082201
ENSDART00000189448 |
slc22a13b
|
solute carrier family 22 member 13b |
| chr8_+_28259347 | 0.31 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr1_+_34527213 | 0.31 |
ENSDART00000180483
|
klf12a
|
Kruppel-like factor 12a |
| chr20_+_5132138 | 0.30 |
ENSDART00000081351
ENSDART00000157685 |
cyp46a1.1
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 1 |
| chr14_-_17306261 | 0.30 |
ENSDART00000191747
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr21_-_26250744 | 0.29 |
ENSDART00000044735
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr4_-_25858620 | 0.28 |
ENSDART00000128968
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr18_+_14693682 | 0.28 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr2_+_51796441 | 0.28 |
ENSDART00000165151
|
crygn1
|
crystallin, gamma N1 |
| chr6_-_28980756 | 0.27 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr21_-_2707768 | 0.26 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr21_+_11105007 | 0.26 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr24_+_15670013 | 0.26 |
ENSDART00000185826
|
CU929414.1
|
|
| chr20_+_1960092 | 0.26 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr21_-_26250443 | 0.25 |
ENSDART00000123713
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr9_+_38292947 | 0.25 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr22_+_9751117 | 0.24 |
ENSDART00000121827
|
BX664625.1
|
|
| chr4_-_25859305 | 0.24 |
ENSDART00000159528
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr1_+_14466860 | 0.23 |
ENSDART00000114097
|
stim2a
|
tromal interaction molecule 2a |
| chr9_-_5263947 | 0.23 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr4_+_76752707 | 0.23 |
ENSDART00000082121
|
ms4a17a.4
|
membrane-spanning 4-domains, subfamily A, member 17A.4 |
| chr1_-_12161148 | 0.23 |
ENSDART00000166429
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr7_-_36358735 | 0.22 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr10_-_5821584 | 0.22 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr15_-_25527580 | 0.22 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr14_-_21959712 | 0.21 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr11_-_27702778 | 0.21 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr16_+_49005321 | 0.20 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr2_-_732864 | 0.19 |
ENSDART00000168585
|
CU861459.1
|
|
| chr19_+_27342479 | 0.19 |
ENSDART00000184687
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr13_-_18195942 | 0.19 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr6_-_49159207 | 0.18 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr16_-_44709832 | 0.18 |
ENSDART00000156784
|
si:ch211-151m7.6
|
si:ch211-151m7.6 |
| chr23_-_36934944 | 0.18 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr13_+_45772924 | 0.17 |
ENSDART00000146964
|
si:ch211-62a1.3
|
si:ch211-62a1.3 |
| chr4_-_73266929 | 0.17 |
ENSDART00000158194
|
wu:fc30c06
|
wu:fc30c06 |
| chr18_+_31410652 | 0.17 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr24_+_2962988 | 0.17 |
ENSDART00000164449
|
eci2
|
enoyl-CoA delta isomerase 2 |
| chr25_+_22643954 | 0.17 |
ENSDART00000182843
ENSDART00000121791 |
ush1c
|
Usher syndrome 1C |
| chr2_-_144393 | 0.16 |
ENSDART00000156008
|
adcy1b
|
adenylate cyclase 1b |
| chr9_-_19583704 | 0.16 |
ENSDART00000053218
|
tmprss3a
|
transmembrane protease, serine 3a |
| chr18_-_48755037 | 0.16 |
ENSDART00000109673
|
tirap
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr1_-_10347198 | 0.15 |
ENSDART00000109141
ENSDART00000152565 |
si:ch211-243g6.3
|
si:ch211-243g6.3 |
| chr8_+_18615938 | 0.14 |
ENSDART00000089161
|
si:ch211-232d19.4
|
si:ch211-232d19.4 |
| chr4_-_13902188 | 0.14 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr14_-_2318590 | 0.14 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr12_+_7254112 | 0.14 |
ENSDART00000092009
|
tfam
|
transcription factor A, mitochondrial |
| chr3_-_22214029 | 0.14 |
ENSDART00000154818
|
maptb
|
microtubule-associated protein tau b |
| chr12_-_21834611 | 0.14 |
ENSDART00000179553
|
thrab
|
thyroid hormone receptor alpha b |
| chr20_+_34596205 | 0.14 |
ENSDART00000138338
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr21_+_35289469 | 0.13 |
ENSDART00000155140
|
il12bb
|
interleukin 12B, b |
| chr15_-_669476 | 0.13 |
ENSDART00000153687
ENSDART00000030603 |
si:ch211-210b2.2
|
si:ch211-210b2.2 |
| chr23_+_45027263 | 0.12 |
ENSDART00000058364
|
hmgb2b
|
high mobility group box 2b |
| chr9_+_41156818 | 0.12 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr19_+_206835 | 0.12 |
ENSDART00000161137
|
scn1bb
|
sodium channel, voltage-gated, type I, beta b |
| chr2_-_9607879 | 0.12 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr5_+_36661058 | 0.12 |
ENSDART00000125653
|
capns1a
|
calpain, small subunit 1 a |
| chr5_-_54160809 | 0.12 |
ENSDART00000158981
|
arhgef15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr13_-_47810394 | 0.12 |
ENSDART00000105025
|
zmp:0000001138
|
zmp:0000001138 |
| chr17_+_23255365 | 0.11 |
ENSDART00000180277
|
AL935174.5
|
|
| chr24_-_25788841 | 0.11 |
ENSDART00000132235
|
klhl24b
|
kelch-like family member 24b |
| chr20_+_12702923 | 0.10 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr14_-_8625845 | 0.10 |
ENSDART00000054760
|
zgc:162144
|
zgc:162144 |
| chr2_+_53357953 | 0.10 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr8_-_13184989 | 0.10 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr18_+_19842569 | 0.10 |
ENSDART00000147470
|
iqch
|
IQ motif containing H |
| chr1_-_55888970 | 0.09 |
ENSDART00000064194
|
asf1bb
|
anti-silencing function 1Bb histone chaperone |
| chr21_-_280769 | 0.09 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr7_-_44970682 | 0.09 |
ENSDART00000144591
|
fam96b
|
family with sequence similarity 96, member B |
| chr20_-_37287107 | 0.08 |
ENSDART00000076309
|
nmbr
|
neuromedin B receptor |
| chr6_-_32315494 | 0.08 |
ENSDART00000127879
|
atg4c
|
autophagy related 4C, cysteine peptidase |
| chr15_+_11840311 | 0.08 |
ENSDART00000167671
|
prkd2
|
protein kinase D2 |
| chr1_-_19233890 | 0.08 |
ENSDART00000127145
|
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr15_+_15390882 | 0.08 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr15_-_39955785 | 0.07 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr16_-_4769877 | 0.07 |
ENSDART00000149421
ENSDART00000054078 |
rpa2
|
replication protein A2 |
| chr17_+_24722646 | 0.07 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr10_-_39198942 | 0.07 |
ENSDART00000077619
|
b3gat1b
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) b |
| chr21_-_35428039 | 0.07 |
ENSDART00000193298
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr3_-_12227359 | 0.07 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr21_+_6197223 | 0.06 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr23_+_41679586 | 0.06 |
ENSDART00000067662
|
CU914487.1
|
|
| chr10_+_44903676 | 0.06 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr1_+_27713380 | 0.06 |
ENSDART00000169584
|
si:dkey-29d5.1
|
si:dkey-29d5.1 |
| chr23_+_37955041 | 0.05 |
ENSDART00000087148
|
cbln4
|
cerebellin 4 precursor |
| chr17_+_25331576 | 0.05 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr5_-_30079434 | 0.05 |
ENSDART00000133981
|
bco2a
|
beta-carotene oxygenase 2a |
| chr12_-_43428542 | 0.05 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr18_-_19350792 | 0.05 |
ENSDART00000147902
|
megf11
|
multiple EGF-like-domains 11 |
| chr6_+_11026866 | 0.05 |
ENSDART00000189619
|
abcc6a
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6a |
| chr12_-_16380961 | 0.05 |
ENSDART00000086984
|
hectd2
|
HECT domain containing 2 |
| chr9_-_54304684 | 0.04 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr8_-_38810233 | 0.04 |
ENSDART00000085304
|
pcsk5b
|
proprotein convertase subtilisin/kexin type 5b |
| chr5_-_57526807 | 0.04 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr18_-_17724295 | 0.03 |
ENSDART00000121553
|
fam192a
|
family with sequence similarity 192, member A |
| chr1_+_9086942 | 0.03 |
ENSDART00000055023
|
cacng3a
|
calcium channel, voltage-dependent, gamma subunit 3a |
| chr17_+_28882977 | 0.03 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr11_+_37612672 | 0.03 |
ENSDART00000157993
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr1_+_23408622 | 0.03 |
ENSDART00000140706
|
chrna9
|
cholinergic receptor, nicotinic, alpha 9 |
| chr17_-_15611744 | 0.03 |
ENSDART00000010496
|
fhl5
|
four and a half LIM domains 5 |
| chr2_+_45521883 | 0.03 |
ENSDART00000139580
|
aknad1
|
AKNA domain containing 1 |
| chr18_+_16133595 | 0.03 |
ENSDART00000080423
|
ctsd
|
cathepsin D |
| chr15_-_31419805 | 0.03 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr5_+_37890521 | 0.03 |
ENSDART00000140207
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr2_+_3881000 | 0.02 |
ENSDART00000081897
|
mpp7b
|
membrane protein, palmitoylated 7b (MAGUK p55 subfamily member 7) |
| chr23_-_7826849 | 0.02 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr6_-_43882696 | 0.02 |
ENSDART00000064938
|
foxp1b
|
forkhead box P1b |
| chr2_+_1881334 | 0.02 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr22_-_12862415 | 0.02 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr8_-_8698607 | 0.02 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr24_+_13017586 | 0.02 |
ENSDART00000142457
|
stau2
|
staufen double-stranded RNA binding protein 2 |
| chr15_-_762319 | 0.02 |
ENSDART00000154306
ENSDART00000157492 |
si:dkey-7i4.16
znf1011
|
si:dkey-7i4.16 zinc finger protein 1011 |
| chr3_-_39305291 | 0.02 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr24_-_12981423 | 0.01 |
ENSDART00000133166
ENSDART00000131370 ENSDART00000002992 |
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr19_+_35002479 | 0.01 |
ENSDART00000103266
|
wdyhv1
|
WDYHV motif containing 1 |
| chr3_-_18711288 | 0.01 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr20_-_36393555 | 0.01 |
ENSDART00000153421
|
si:dkey-1j5.4
|
si:dkey-1j5.4 |
| chr22_+_2247645 | 0.01 |
ENSDART00000143366
ENSDART00000147852 |
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr8_-_37461835 | 0.01 |
ENSDART00000013446
|
eif2d
|
eukaryotic translation initiation factor 2D |
| chr4_-_73572030 | 0.01 |
ENSDART00000121652
|
znf1015
|
zinc finger protein 1015 |
Network of associatons between targets according to the STRING database.
First level regulatory network of six6a+six6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.2 | 1.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.2 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.4 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.4 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.9 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 2.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.9 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.7 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.0 | 0.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 2.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 1.2 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.2 | GO:0046549 | startle response(GO:0001964) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 1.4 | GO:0006979 | response to oxidative stress(GO:0006979) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.4 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.8 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 2.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 1.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 1.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.6 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.3 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 1.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.3 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.5 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 1.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |