Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
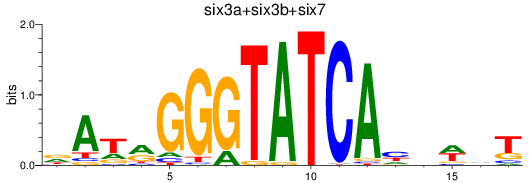
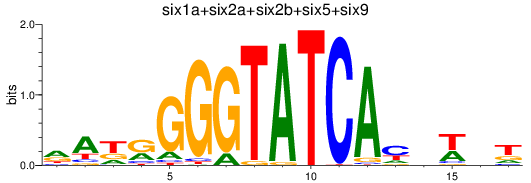
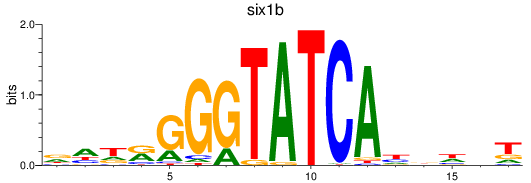
Results for six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
Z-value: 1.50
Transcription factors associated with six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six3b
|
ENSDARG00000054879 | SIX homeobox 3b |
|
six3a
|
ENSDARG00000058008 | SIX homeobox 3a |
|
six7
|
ENSDARG00000070107 | SIX homeobox 7 |
|
six3a
|
ENSDARG00000114971 | SIX homeobox 3a |
|
six1a
|
ENSDARG00000039304 | SIX homeobox 1a |
|
six2b
|
ENSDARG00000054878 | SIX homeobox 2b |
|
six2a
|
ENSDARG00000058004 | SIX homeobox 2a |
|
six5
|
ENSDARG00000068406 | SIX homeobox 5 |
|
six9
|
ENSDARG00000068407 | SIX homeobox 9 |
|
six2a
|
ENSDARG00000115214 | SIX homeobox 2a |
|
six1b
|
ENSDARG00000026473 | SIX homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six5 | dr11_v1_chr18_-_37252036_37252036 | 0.93 | 3.4e-04 | Click! |
| six3b | dr11_v1_chr12_+_25600685_25600685 | -0.91 | 5.8e-04 | Click! |
| six1b | dr11_v1_chr20_+_20499869_20499869 | -0.91 | 6.2e-04 | Click! |
| six2b | dr11_v1_chr12_-_25612170_25612170 | -0.91 | 7.3e-04 | Click! |
| six1a | dr11_v1_chr13_-_31622195_31622195 | -0.88 | 1.8e-03 | Click! |
| six2a | dr11_v1_chr13_+_10232695_10232695 | -0.86 | 3.1e-03 | Click! |
| six3a | dr11_v1_chr13_-_10261383_10261383 | -0.82 | 7.0e-03 | Click! |
| six9 | dr11_v1_chr18_-_37241080_37241115 | -0.55 | 1.3e-01 | Click! |
| six7 | dr11_v1_chr7_-_7420301_7420301 | -0.35 | 3.6e-01 | Click! |
Activity profile of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b motif
Sorted Z-values of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_42898220 | 5.45 |
ENSDART00000099270
|
CU326366.2
|
|
| chr19_-_20403845 | 4.30 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr1_-_24255149 | 3.01 |
ENSDART00000146960
|
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr21_-_7940043 | 2.99 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr10_+_10972795 | 2.86 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr15_-_30505607 | 2.60 |
ENSDART00000155212
|
msi2b
|
musashi RNA-binding protein 2b |
| chr25_-_21763750 | 2.50 |
ENSDART00000089596
|
tmem168b
|
transmembrane protein 168b |
| chr16_+_50755133 | 2.48 |
ENSDART00000029283
|
IGLON5
|
zgc:110372 |
| chr15_+_17446796 | 2.22 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr20_+_37866861 | 2.15 |
ENSDART00000153220
|
vash2
|
vasohibin 2 |
| chr10_+_42898103 | 2.09 |
ENSDART00000015872
|
zcchc9
|
zinc finger, CCHC domain containing 9 |
| chr19_+_34230108 | 2.07 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr17_+_16429826 | 1.78 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr23_+_39481537 | 1.72 |
ENSDART00000187222
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr16_-_46567344 | 1.72 |
ENSDART00000127721
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr16_+_26735894 | 1.70 |
ENSDART00000191605
ENSDART00000185175 |
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr16_+_41060161 | 1.67 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr20_+_4221978 | 1.60 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr14_+_22397251 | 1.56 |
ENSDART00000185239
ENSDART00000124072 ENSDART00000054977 |
atp7a
|
ATPase copper transporting alpha |
| chr12_+_13742953 | 1.54 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr21_-_21465111 | 1.45 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr17_-_2573021 | 1.40 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr12_+_13742778 | 1.32 |
ENSDART00000111401
ENSDART00000190552 |
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr20_+_22067337 | 1.22 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr21_-_26028205 | 1.16 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr9_-_13871935 | 1.08 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr8_-_25566347 | 1.06 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr7_-_71486162 | 1.06 |
ENSDART00000045253
|
aco1
|
aconitase 1, soluble |
| chr17_-_6600899 | 1.06 |
ENSDART00000154074
ENSDART00000180912 |
ANKRD66
|
si:ch211-189e2.2 |
| chr8_+_20440297 | 1.05 |
ENSDART00000189486
|
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr4_-_7811925 | 1.04 |
ENSDART00000103070
|
cdk17
|
cyclin-dependent kinase 17 |
| chr20_+_34502606 | 1.02 |
ENSDART00000139739
|
gorab
|
golgin, rab6-interacting |
| chr11_-_24016761 | 1.00 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr18_-_49283058 | 0.98 |
ENSDART00000076554
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr5_+_37903790 | 0.92 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr5_-_26247973 | 0.90 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr21_+_18405585 | 0.87 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr5_+_41477526 | 0.87 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr21_+_45519373 | 0.85 |
ENSDART00000183665
|
fnip1
|
folliculin interacting protein 1 |
| chr5_+_41477954 | 0.84 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr7_-_29356084 | 0.73 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr23_-_27045231 | 0.70 |
ENSDART00000187979
|
zgc:66440
|
zgc:66440 |
| chr13_+_33368140 | 0.68 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr13_-_3324764 | 0.65 |
ENSDART00000102748
ENSDART00000114040 |
ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr17_+_13099476 | 0.65 |
ENSDART00000012670
|
pnn
|
pinin, desmosome associated protein |
| chr1_+_532766 | 0.63 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr13_+_25549425 | 0.61 |
ENSDART00000087553
ENSDART00000169199 |
sec23ip
|
SEC23 interacting protein |
| chr2_-_32366287 | 0.59 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr5_-_54395488 | 0.57 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr7_-_59515569 | 0.57 |
ENSDART00000163343
ENSDART00000165457 ENSDART00000163745 |
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr6_-_7686594 | 0.56 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr21_-_13972745 | 0.55 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr12_+_9183626 | 0.55 |
ENSDART00000020192
|
tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr24_+_30435164 | 0.54 |
ENSDART00000193877
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr2_-_27473645 | 0.53 |
ENSDART00000186152
|
CU469530.2
|
|
| chr20_+_4222357 | 0.53 |
ENSDART00000188331
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr13_-_18122333 | 0.53 |
ENSDART00000128748
|
washc2c
|
WASH complex subunit 2C |
| chr9_-_28990649 | 0.52 |
ENSDART00000078823
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr22_-_5171362 | 0.51 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr8_-_26961779 | 0.50 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr8_+_7801060 | 0.49 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr3_-_5067585 | 0.48 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr20_-_45058041 | 0.48 |
ENSDART00000124298
|
klhl29
|
kelch-like family member 29 |
| chr5_+_13326765 | 0.43 |
ENSDART00000090851
|
ydjc
|
YdjC chitooligosaccharide deacetylase homolog |
| chr25_-_37180969 | 0.41 |
ENSDART00000152338
|
tdrd12
|
tudor domain containing 12 |
| chr20_+_13783040 | 0.41 |
ENSDART00000115329
ENSDART00000152497 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr11_+_1602916 | 0.39 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr6_-_40899618 | 0.38 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr10_-_32610776 | 0.36 |
ENSDART00000017436
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr15_-_43270889 | 0.35 |
ENSDART00000166805
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr14_-_470505 | 0.35 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr15_-_38218320 | 0.35 |
ENSDART00000152363
|
rhoga
|
ras homolog family member Ga |
| chr1_+_37752171 | 0.34 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr24_-_18809433 | 0.33 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr15_+_40792667 | 0.33 |
ENSDART00000193186
ENSDART00000184559 |
fat3a
|
FAT atypical cadherin 3a |
| chr10_+_14488625 | 0.32 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr7_+_56517402 | 0.30 |
ENSDART00000073597
|
dhodh
|
dihydroorotate dehydrogenase |
| chr2_+_6999369 | 0.30 |
ENSDART00000050597
|
xpr1b
|
xenotropic and polytropic retrovirus receptor 1b |
| chr7_-_38183331 | 0.29 |
ENSDART00000149382
|
abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr2_-_38312757 | 0.29 |
ENSDART00000167274
|
ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr4_+_20085114 | 0.27 |
ENSDART00000186698
ENSDART00000188635 |
ppp6r2a
|
protein phosphatase 6, regulatory subunit 2a |
| chr2_+_26240631 | 0.26 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr15_+_21882419 | 0.26 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr13_-_40120252 | 0.25 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr6_-_28222592 | 0.25 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr5_+_26806141 | 0.24 |
ENSDART00000142753
|
rabl6b
|
RAB, member RAS oncogene family-like 6b |
| chr15_-_15910226 | 0.23 |
ENSDART00000154219
|
synrg
|
synergin, gamma |
| chr19_-_22664128 | 0.23 |
ENSDART00000136295
|
eif3eb
|
eukaryotic translation initiation factor 3, subunit E, b |
| chr11_+_24002503 | 0.23 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr10_+_40624702 | 0.22 |
ENSDART00000109129
|
zgc:172131
|
zgc:172131 |
| chr11_-_44194132 | 0.22 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr17_+_31739418 | 0.22 |
ENSDART00000155073
ENSDART00000156180 |
arhgap5
|
Rho GTPase activating protein 5 |
| chr4_-_49720388 | 0.22 |
ENSDART00000154146
|
znf1057
|
zinc finger protein 1057 |
| chr17_+_10593398 | 0.19 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr7_-_40657831 | 0.19 |
ENSDART00000084153
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr20_-_44460789 | 0.19 |
ENSDART00000085442
|
mut
|
methylmalonyl CoA mutase |
| chr14_+_1355857 | 0.18 |
ENSDART00000188008
|
bbs12
|
Bardet-Biedl syndrome 12 |
| chr22_-_24967348 | 0.18 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr22_-_11520405 | 0.17 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr25_-_20238793 | 0.16 |
ENSDART00000145987
|
dnm1l
|
dynamin 1-like |
| chr3_-_21094437 | 0.16 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr25_-_19068557 | 0.14 |
ENSDART00000184780
|
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr17_+_132555 | 0.14 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr9_+_46644633 | 0.14 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr9_+_26086135 | 0.14 |
ENSDART00000011910
|
arglu1a
|
arginine and glutamate rich 1a |
| chr22_+_2315996 | 0.13 |
ENSDART00000132489
|
znf1175
|
zinc finger protein 1175 |
| chr10_+_35468978 | 0.13 |
ENSDART00000131984
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr4_-_13502549 | 0.12 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr21_-_2158298 | 0.11 |
ENSDART00000182199
|
gb:ai877918
|
expressed sequence AI877918 |
| chr6_+_52947699 | 0.11 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr19_-_11949996 | 0.11 |
ENSDART00000163478
ENSDART00000167299 |
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr7_-_47381146 | 0.11 |
ENSDART00000185596
|
AL935126.2
|
|
| chr25_+_18953756 | 0.11 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr7_-_36113098 | 0.11 |
ENSDART00000064913
|
fto
|
fat mass and obesity associated |
| chr9_+_53637932 | 0.11 |
ENSDART00000188962
|
CABZ01084081.1
|
|
| chr15_-_945804 | 0.10 |
ENSDART00000063257
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr6_+_41186320 | 0.09 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr6_+_52947186 | 0.09 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr13_+_9678427 | 0.08 |
ENSDART00000153907
|
sema4ga
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ga |
| chr5_-_12823876 | 0.06 |
ENSDART00000019461
|
p2rx2
|
purinergic receptor P2X, ligand-gated ion channel, 2 |
| chr4_+_19127973 | 0.06 |
ENSDART00000136611
|
si:dkey-21o22.2
|
si:dkey-21o22.2 |
| chr19_-_15434813 | 0.06 |
ENSDART00000019843
|
ftr55
|
finTRIM family, member 55 |
| chr7_-_72423666 | 0.05 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr25_+_3788074 | 0.05 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr13_+_9678262 | 0.05 |
ENSDART00000111365
|
sema4ga
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ga |
| chr2_-_51275873 | 0.04 |
ENSDART00000168019
|
si:ch211-215e19.4
|
si:ch211-215e19.4 |
| chr10_+_40568735 | 0.04 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr16_-_46567136 | 0.03 |
ENSDART00000159180
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr23_+_3721042 | 0.03 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr11_+_24339377 | 0.03 |
ENSDART00000133679
ENSDART00000135435 ENSDART00000017973 ENSDART00000131365 ENSDART00000186418 |
rbm39a
|
RNA binding motif protein 39a |
| chr9_+_35017702 | 0.01 |
ENSDART00000193640
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr11_-_42750626 | 0.00 |
ENSDART00000130640
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr8_+_23745167 | 0.00 |
ENSDART00000149352
|
si:ch211-163l21.11
|
si:ch211-163l21.11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.6 | 1.7 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.4 | 3.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.3 | 1.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 1.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.2 | 2.6 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.7 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.2 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.5 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 2.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 3.1 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.1 | 2.9 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.3 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 1.4 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 1.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.3 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.0 | 1.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.2 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 1.1 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 1.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 2.1 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.2 | 0.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.6 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.7 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 2.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.5 | GO:0043296 | apical junction complex(GO:0043296) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.5 | 4.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 3.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 1.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.3 | 1.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 1.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 1.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.5 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.3 | GO:0015562 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.7 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 1.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 1.7 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 3.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.4 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |