Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for si:dkey-18a10.3
Z-value: 1.50
Transcription factors associated with si:dkey-18a10.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-18a10.3
|
ENSDARG00000090814 | si_dkey-18a10.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-18a10.3 | dr11_v1_chr10_-_33572441_33572441 | 0.68 | 4.2e-02 | Click! |
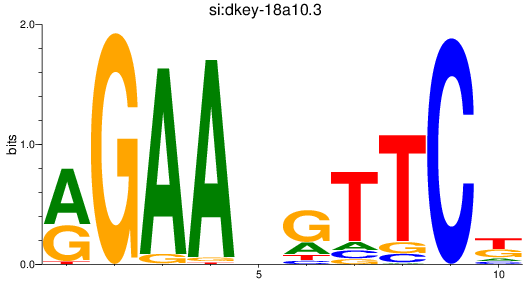
Activity profile of si:dkey-18a10.3 motif
Sorted Z-values of si:dkey-18a10.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_31372639 | 3.94 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr9_-_22318511 | 3.64 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr20_-_53996193 | 3.64 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr9_-_11560427 | 3.64 |
ENSDART00000127942
ENSDART00000061442 |
cryba2b
|
crystallin, beta A2b |
| chr7_+_38750871 | 2.71 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr7_+_29954709 | 2.71 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr9_-_22299412 | 2.68 |
ENSDART00000139101
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr21_+_20383837 | 2.51 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr20_-_44496245 | 2.44 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr7_+_29955368 | 2.42 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr20_-_49657134 | 2.36 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr16_+_34523515 | 2.33 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr15_+_36941490 | 2.10 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr19_+_19767567 | 2.09 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr2_-_35566938 | 2.04 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr17_+_41463942 | 2.01 |
ENSDART00000075331
|
insm1b
|
insulinoma-associated 1b |
| chr13_-_31452516 | 2.00 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr22_-_14115292 | 1.99 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr6_+_49095646 | 1.95 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr15_-_452347 | 1.95 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr7_-_51102479 | 1.86 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr20_+_15552657 | 1.85 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr9_-_1951144 | 1.80 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr13_+_24834199 | 1.75 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr7_-_60831082 | 1.71 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr14_-_1990290 | 1.63 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr4_-_16353733 | 1.58 |
ENSDART00000186785
|
lum
|
lumican |
| chr9_-_56272465 | 1.56 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_-_58650057 | 1.55 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr1_-_59411901 | 1.51 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr13_-_23007813 | 1.51 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr9_-_22023061 | 1.50 |
ENSDART00000101952
|
crygm2c
|
crystallin, gamma M2c |
| chr25_+_28272908 | 1.47 |
ENSDART00000010325
|
fezf1
|
FEZ family zinc finger 1 |
| chr10_+_18952271 | 1.42 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr9_+_44431174 | 1.39 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr15_-_37850969 | 1.38 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr10_+_21776911 | 1.38 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr23_+_36178104 | 1.37 |
ENSDART00000103131
|
hoxc1a
|
homeobox C1a |
| chr14_+_36223097 | 1.37 |
ENSDART00000186872
|
pitx2
|
paired-like homeodomain 2 |
| chr4_-_17055782 | 1.34 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr6_-_60147517 | 1.29 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr6_+_16406723 | 1.23 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr5_+_32791245 | 1.22 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr23_-_24682244 | 1.18 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr17_-_42213285 | 1.17 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr11_+_2180072 | 1.16 |
ENSDART00000186854
ENSDART00000164839 |
hoxc11b
|
homeobox C11b |
| chr4_-_17055950 | 1.14 |
ENSDART00000162945
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr9_+_32301017 | 1.13 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr19_-_35596207 | 1.09 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr19_-_9712530 | 1.09 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr8_+_22931427 | 1.08 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr21_-_25756119 | 1.08 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr9_+_44430974 | 0.99 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr9_+_32301456 | 0.98 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr25_+_19999623 | 0.97 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr6_+_202367 | 0.97 |
ENSDART00000168232
|
mgat3b
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase b |
| chr1_+_46652615 | 0.97 |
ENSDART00000053221
|
arl11
|
ADP-ribosylation factor-like 11 |
| chr6_-_40286633 | 0.96 |
ENSDART00000033844
|
col7a1
|
collagen, type VII, alpha 1 |
| chr19_+_35799384 | 0.94 |
ENSDART00000076023
|
angpt2b
|
angiopoietin 2b |
| chr6_+_9175886 | 0.92 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr3_-_53722182 | 0.90 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr17_+_21964472 | 0.89 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr10_+_17875891 | 0.88 |
ENSDART00000191744
|
phf24
|
PHD finger protein 24 |
| chr4_-_24019711 | 0.83 |
ENSDART00000077926
|
celf2
|
cugbp, Elav-like family member 2 |
| chr15_-_22147860 | 0.83 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr5_-_23999777 | 0.81 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr24_-_21808816 | 0.81 |
ENSDART00000025621
ENSDART00000130446 |
flt1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) |
| chr6_-_57655299 | 0.81 |
ENSDART00000083807
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr18_+_8346920 | 0.78 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr13_-_45475289 | 0.78 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr8_-_1909840 | 0.77 |
ENSDART00000147408
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr10_-_26273629 | 0.76 |
ENSDART00000147790
|
dchs1b
|
dachsous cadherin-related 1b |
| chr11_+_10548171 | 0.74 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr16_+_21330634 | 0.74 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr19_+_12237945 | 0.73 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr6_-_13401133 | 0.73 |
ENSDART00000151206
|
fmnl2b
|
formin-like 2b |
| chr22_+_24389135 | 0.72 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr12_+_19866865 | 0.70 |
ENSDART00000045609
|
cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr9_-_327901 | 0.70 |
ENSDART00000159956
|
ndufa4l2a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2a |
| chr22_+_22021936 | 0.69 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr6_-_57655030 | 0.69 |
ENSDART00000155438
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr25_-_18470695 | 0.69 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr21_-_5881344 | 0.67 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr4_-_43822179 | 0.67 |
ENSDART00000185246
|
si:dkey-261p22.1
|
si:dkey-261p22.1 |
| chr6_+_13333133 | 0.66 |
ENSDART00000038505
|
rprmb
|
reprimo, TP53 dependent G2 arrest mediator candidate b |
| chr18_+_38321039 | 0.65 |
ENSDART00000132534
ENSDART00000111260 ENSDART00000192806 |
alx4b
|
ALX homeobox 4b |
| chr18_+_14477740 | 0.65 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr20_+_34543365 | 0.65 |
ENSDART00000152073
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr23_-_44494401 | 0.62 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr1_+_2101541 | 0.61 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr3_+_26267725 | 0.61 |
ENSDART00000131288
|
adap2
|
ArfGAP with dual PH domains 2 |
| chr5_+_20103085 | 0.60 |
ENSDART00000079424
|
si:rp71-1c23.3
|
si:rp71-1c23.3 |
| chr10_-_36682509 | 0.60 |
ENSDART00000148093
ENSDART00000063365 |
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr15_+_46329149 | 0.59 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr6_+_46341306 | 0.59 |
ENSDART00000111905
|
BX649498.1
|
|
| chr9_+_44430705 | 0.59 |
ENSDART00000190696
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr4_+_35553514 | 0.57 |
ENSDART00000182938
|
CR847906.1
|
|
| chr23_-_6879731 | 0.56 |
ENSDART00000082154
|
trim35-39
|
tripartite motif containing 35-39 |
| chr1_+_31054915 | 0.56 |
ENSDART00000148968
|
itga6b
|
integrin, alpha 6b |
| chr25_-_17403598 | 0.56 |
ENSDART00000104216
|
cyp2x9
|
cytochrome P450, family 2, subfamily X, polypeptide 9 |
| chr9_+_53707240 | 0.55 |
ENSDART00000171490
|
PCDH8
|
si:ch211-199f5.1 |
| chr3_-_36612877 | 0.55 |
ENSDART00000167164
|
si:dkeyp-72e1.7
|
si:dkeyp-72e1.7 |
| chr3_+_24634481 | 0.52 |
ENSDART00000163080
|
si:dkey-68o6.8
|
si:dkey-68o6.8 |
| chr4_+_8638622 | 0.52 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr8_+_650342 | 0.52 |
ENSDART00000048498
|
LO018102.1
|
|
| chr14_+_48045193 | 0.52 |
ENSDART00000124773
|
ppid
|
peptidylprolyl isomerase D |
| chr7_+_17229282 | 0.52 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr15_-_4027367 | 0.52 |
ENSDART00000179889
|
CR626884.5
|
|
| chr17_+_729423 | 0.52 |
ENSDART00000164795
|
slc25a47b
|
solute carrier family 25, member 47b |
| chr5_-_30176970 | 0.51 |
ENSDART00000098300
|
adamts8a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8a |
| chr25_+_35250976 | 0.51 |
ENSDART00000003494
|
slc17a6a
|
solute carrier family 17 (vesicular glutamate transporter), member 6a |
| chr24_+_21973929 | 0.50 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr9_+_6587056 | 0.50 |
ENSDART00000193421
|
fhl2a
|
four and a half LIM domains 2a |
| chr19_-_3193912 | 0.49 |
ENSDART00000133159
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr1_-_681116 | 0.49 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr8_+_25342896 | 0.47 |
ENSDART00000129032
|
CR847543.1
|
|
| chr1_-_9644630 | 0.47 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr10_-_26274094 | 0.46 |
ENSDART00000108798
|
dchs1b
|
dachsous cadherin-related 1b |
| chr8_-_13013123 | 0.46 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr1_-_53714885 | 0.45 |
ENSDART00000026409
|
cct4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr1_-_1631399 | 0.45 |
ENSDART00000176787
|
clic6
|
chloride intracellular channel 6 |
| chr20_+_37393134 | 0.44 |
ENSDART00000128321
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr9_+_6587364 | 0.44 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr18_-_28938912 | 0.44 |
ENSDART00000136201
|
si:ch211-174j14.2
|
si:ch211-174j14.2 |
| chr15_-_24883956 | 0.43 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr17_-_44584811 | 0.43 |
ENSDART00000165059
ENSDART00000165252 |
slc35f4
|
solute carrier family 35, member F4 |
| chr13_+_18202703 | 0.42 |
ENSDART00000109642
|
tet1
|
tet methylcytosine dioxygenase 1 |
| chr25_-_14433503 | 0.40 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr2_-_35103069 | 0.39 |
ENSDART00000111730
|
pappa2
|
pappalysin 2 |
| chr20_-_27864964 | 0.38 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr23_+_45611980 | 0.37 |
ENSDART00000181582
|
dclk2b
|
doublecortin-like kinase 2b |
| chr18_-_45617146 | 0.37 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr6_-_40697585 | 0.37 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr11_-_5939861 | 0.36 |
ENSDART00000110033
|
abhd8b
|
abhydrolase domain containing 8b |
| chr10_+_19039507 | 0.35 |
ENSDART00000193538
ENSDART00000111952 ENSDART00000180093 |
igsf9a
|
immunoglobulin superfamily, member 9a |
| chr13_+_22865676 | 0.34 |
ENSDART00000057651
|
hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr11_-_23501467 | 0.34 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr4_-_52165969 | 0.33 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr18_+_15758375 | 0.33 |
ENSDART00000137554
|
si:ch211-219a15.4
|
si:ch211-219a15.4 |
| chr11_-_5939460 | 0.33 |
ENSDART00000181474
|
abhd8b
|
abhydrolase domain containing 8b |
| chr8_+_52637507 | 0.31 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr2_-_41620112 | 0.30 |
ENSDART00000138822
ENSDART00000004816 |
zgc:110158
|
zgc:110158 |
| chr21_+_27297145 | 0.30 |
ENSDART00000146588
|
si:dkey-175m17.6
|
si:dkey-175m17.6 |
| chr23_-_39738055 | 0.29 |
ENSDART00000137859
|
htr6
|
5-hydroxytryptamine (serotonin) receptor 6 |
| chr21_+_21713531 | 0.29 |
ENSDART00000140501
|
or123-1
|
odorant receptor, family E, subfamily 123, member 1 |
| chr9_+_54290896 | 0.29 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr22_-_607812 | 0.28 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr4_+_8168514 | 0.27 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr22_+_18886209 | 0.27 |
ENSDART00000144402
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr24_+_20969358 | 0.26 |
ENSDART00000129675
|
drd3
|
dopamine receptor D3 |
| chr25_+_14165447 | 0.25 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr14_-_17622080 | 0.25 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr16_+_45922175 | 0.25 |
ENSDART00000018253
|
rbm8a
|
RNA binding motif protein 8A |
| chr22_-_4742866 | 0.25 |
ENSDART00000177992
|
CR751224.1
|
|
| chr20_-_8304271 | 0.24 |
ENSDART00000179057
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr12_+_45200744 | 0.24 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr6_+_21684296 | 0.23 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr19_-_3193443 | 0.23 |
ENSDART00000179855
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr13_+_25486608 | 0.23 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr14_-_2318590 | 0.21 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr12_-_37941733 | 0.21 |
ENSDART00000130167
|
CABZ01046949.1
|
|
| chr2_+_24199073 | 0.21 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr3_+_33345348 | 0.21 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr8_+_14778292 | 0.20 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr16_-_13662514 | 0.20 |
ENSDART00000146348
|
shisa7a
|
shisa family member 7a |
| chr2_+_24199276 | 0.20 |
ENSDART00000140575
|
map4l
|
microtubule associated protein 4 like |
| chr18_+_50907675 | 0.20 |
ENSDART00000159950
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr18_+_3338228 | 0.19 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr21_-_2168515 | 0.19 |
ENSDART00000165455
|
BX510922.2
|
|
| chr14_+_22022441 | 0.18 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr3_-_23575007 | 0.17 |
ENSDART00000155282
ENSDART00000087726 |
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr23_+_24931999 | 0.17 |
ENSDART00000136162
ENSDART00000140335 |
klhl21
|
kelch-like family member 21 |
| chr8_+_28467893 | 0.17 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr10_+_34426571 | 0.17 |
ENSDART00000144529
|
nbeaa
|
neurobeachin a |
| chr10_-_36738619 | 0.17 |
ENSDART00000093000
ENSDART00000157179 |
PLEKHB1
|
si:ch211-176g13.7 |
| chr1_-_47431453 | 0.16 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr7_-_71837213 | 0.16 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr5_-_30475011 | 0.16 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr2_-_9260002 | 0.15 |
ENSDART00000057299
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr17_-_31044803 | 0.15 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr3_+_6443992 | 0.14 |
ENSDART00000169325
ENSDART00000162255 |
nup85
|
nucleoporin 85 |
| chr3_+_1015867 | 0.14 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr3_+_1037946 | 0.14 |
ENSDART00000167590
ENSDART00000011111 |
zgc:153921
zgc:153921
|
zgc:153921 zgc:153921 |
| chr16_+_46666074 | 0.14 |
ENSDART00000074919
ENSDART00000142698 |
ubqln4
|
ubiquilin 4 |
| chr10_+_36349225 | 0.13 |
ENSDART00000174033
ENSDART00000099395 |
or104-2
|
odorant receptor, family C, subfamily 104, member 2 |
| chr17_-_33405301 | 0.13 |
ENSDART00000157089
|
BX323819.1
|
|
| chr18_+_18982077 | 0.12 |
ENSDART00000006300
|
hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr12_-_48312647 | 0.12 |
ENSDART00000114415
|
ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr1_+_54606354 | 0.11 |
ENSDART00000125227
|
si:ch211-202h22.10
|
si:ch211-202h22.10 |
| chr16_-_6821927 | 0.11 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr25_+_35942867 | 0.11 |
ENSDART00000066985
|
hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr7_-_28463106 | 0.10 |
ENSDART00000137799
|
trim66
|
tripartite motif containing 66 |
| chr22_-_27296889 | 0.10 |
ENSDART00000155724
|
si:dkey-208m12.3
|
si:dkey-208m12.3 |
| chr11_+_30000814 | 0.10 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr2_+_31957554 | 0.09 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr5_-_13685047 | 0.09 |
ENSDART00000018351
|
zgc:65851
|
zgc:65851 |
| chr10_+_29850330 | 0.09 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr4_+_33537813 | 0.09 |
ENSDART00000159918
|
si:dkey-84h14.4
|
si:dkey-84h14.4 |
| chr9_+_6802641 | 0.09 |
ENSDART00000187278
|
FO203514.1
|
|
| chr19_+_23301050 | 0.09 |
ENSDART00000193713
|
si:dkey-79i2.4
|
si:dkey-79i2.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-18a10.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.9 | 3.6 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.5 | 2.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.4 | 1.2 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.4 | 1.2 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 1.9 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 2.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 1.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.2 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 0.8 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.2 | 1.9 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 2.5 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 3.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.1 | 2.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.5 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 1.5 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.5 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 2.5 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 2.5 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.5 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 11.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 3.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.4 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 2.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.2 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 1.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.8 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 2.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 4.5 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.8 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 1.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.7 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 2.6 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 3.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.6 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.7 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.6 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 3.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 6.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 3.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.6 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 2.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 3.9 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.6 | 1.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 1.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 1.5 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.7 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 0.7 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 1.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 3.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 11.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 1.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.3 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.1 | 0.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.5 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 3.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 3.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.7 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 4.4 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 3.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 3.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 1.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 1.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |