Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
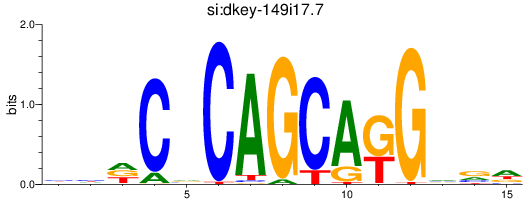
Results for si:dkey-149i17.7_zic1
Z-value: 1.25
Transcription factors associated with si:dkey-149i17.7_zic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-149i17.7
|
ENSDARG00000094684 | si_dkey-149i17.7 |
|
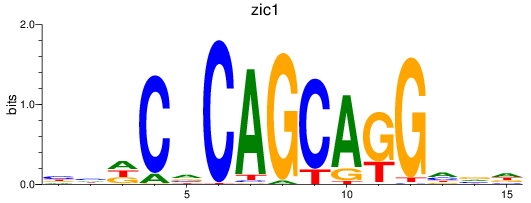
zic1
|
ENSDARG00000015567 | zic family member 1 (odd-paired homolog, Drosophila) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-149i17.7 | dr11_v1_chr2_+_24885987_24885987 | 0.63 | 6.7e-02 | Click! |
| zic1 | dr11_v1_chr24_-_4973765_4973765 | -0.10 | 8.0e-01 | Click! |
Activity profile of si:dkey-149i17.7_zic1 motif
Sorted Z-values of si:dkey-149i17.7_zic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22158784 | 1.34 |
ENSDART00000167850
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr7_-_24699985 | 1.32 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr15_+_9072821 | 1.24 |
ENSDART00000154463
|
si:dkey-202g17.3
|
si:dkey-202g17.3 |
| chr20_-_43775495 | 0.96 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr18_+_907266 | 0.95 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr18_+_49411417 | 0.92 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr24_-_3419998 | 0.82 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr17_-_6382392 | 0.82 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr19_+_43297546 | 0.81 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr22_+_18886209 | 0.80 |
ENSDART00000144402
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr14_-_51855047 | 0.78 |
ENSDART00000088912
|
cplx1
|
complexin 1 |
| chr19_-_32641725 | 0.74 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr17_+_13664442 | 0.70 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr21_+_19445942 | 0.66 |
ENSDART00000030887
|
slc45a2
|
solute carrier family 45, member 2 |
| chr18_+_39487486 | 0.63 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr16_-_55028740 | 0.62 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr1_+_51496862 | 0.61 |
ENSDART00000150433
|
meis1a
|
Meis homeobox 1 a |
| chr4_-_1324141 | 0.61 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr23_+_1349277 | 0.61 |
ENSDART00000173133
ENSDART00000179877 |
utrn
|
utrophin |
| chr19_+_42983613 | 0.61 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr17_+_5915875 | 0.61 |
ENSDART00000184179
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr3_-_55139127 | 0.60 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr3_+_54047342 | 0.58 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr7_+_22543963 | 0.58 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr22_+_5106751 | 0.57 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr3_-_21061931 | 0.57 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr21_+_3093419 | 0.56 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr18_+_7345417 | 0.55 |
ENSDART00000041429
|
glipr1b
|
GLI pathogenesis-related 1b |
| chr16_+_31511739 | 0.53 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr6_+_10338554 | 0.52 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr3_-_18575868 | 0.50 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr4_-_2162688 | 0.50 |
ENSDART00000148900
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr21_+_29077509 | 0.49 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr15_-_16323750 | 0.48 |
ENSDART00000028500
|
nxn
|
nucleoredoxin |
| chr14_-_4043818 | 0.47 |
ENSDART00000179870
|
snx25
|
sorting nexin 25 |
| chr23_+_216012 | 0.47 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr4_-_11024767 | 0.46 |
ENSDART00000067261
ENSDART00000167631 |
tmtc2a
|
transmembrane and tetratricopeptide repeat containing 2a |
| chr25_+_35502552 | 0.46 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr9_+_307863 | 0.45 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr5_-_61588998 | 0.44 |
ENSDART00000050912
|
pex12
|
peroxisomal biogenesis factor 12 |
| chr10_-_22918214 | 0.44 |
ENSDART00000163908
|
rnasekb
|
ribonuclease, RNase K b |
| chr8_+_8845932 | 0.43 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr23_+_35759843 | 0.42 |
ENSDART00000047082
|
gdap1l1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr13_+_30912117 | 0.41 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr20_-_18731268 | 0.41 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr7_-_67842997 | 0.41 |
ENSDART00000169763
|
pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr24_-_30096666 | 0.41 |
ENSDART00000183285
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr3_+_24482999 | 0.41 |
ENSDART00000059179
|
nptxra
|
neuronal pentraxin receptor a |
| chr7_-_13884610 | 0.40 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr21_-_45588720 | 0.40 |
ENSDART00000186642
ENSDART00000189531 |
LO018363.2
|
|
| chr13_-_25632756 | 0.39 |
ENSDART00000077627
ENSDART00000139237 |
ret
|
ret proto-oncogene receptor tyrosine kinase |
| chr16_+_50755133 | 0.38 |
ENSDART00000029283
|
IGLON5
|
zgc:110372 |
| chr10_-_2526526 | 0.38 |
ENSDART00000191726
ENSDART00000192767 |
CU856539.1
|
|
| chr1_+_45085194 | 0.38 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr10_-_25699454 | 0.38 |
ENSDART00000064376
|
sod1
|
superoxide dismutase 1, soluble |
| chr3_+_46479705 | 0.38 |
ENSDART00000181564
|
tyk2
|
tyrosine kinase 2 |
| chr12_+_48634927 | 0.37 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr12_+_4225199 | 0.37 |
ENSDART00000042277
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr25_+_37285737 | 0.37 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr13_+_30912938 | 0.36 |
ENSDART00000190003
|
drgx
|
dorsal root ganglia homeobox |
| chr16_-_27640995 | 0.36 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr19_+_33139164 | 0.36 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr21_+_27448856 | 0.36 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr9_-_23253870 | 0.35 |
ENSDART00000143657
ENSDART00000169911 |
acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr11_+_3254252 | 0.35 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr17_-_52587598 | 0.35 |
ENSDART00000061497
|
si:ch211-173a9.6
|
si:ch211-173a9.6 |
| chr9_-_53920959 | 0.34 |
ENSDART00000149618
|
mid1
|
midline 1 |
| chr3_+_14317802 | 0.34 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr7_+_58843700 | 0.34 |
ENSDART00000159500
ENSDART00000158436 |
lypla1
|
lysophospholipase I |
| chr11_-_669558 | 0.34 |
ENSDART00000173450
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr13_-_11667661 | 0.33 |
ENSDART00000102411
|
dctn1b
|
dynactin 1b |
| chr10_+_37182626 | 0.33 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr25_-_10564721 | 0.33 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr7_-_35126374 | 0.32 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr19_+_310494 | 0.32 |
ENSDART00000053638
|
ctss1
|
cathepsin S, ortholog 1 |
| chr3_+_46479913 | 0.32 |
ENSDART00000149755
|
tyk2
|
tyrosine kinase 2 |
| chr12_+_2665081 | 0.31 |
ENSDART00000147532
|
rbp3
|
retinol binding protein 3 |
| chr5_+_9382301 | 0.31 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr19_+_46259619 | 0.31 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr21_-_40557281 | 0.31 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr5_-_5326010 | 0.30 |
ENSDART00000161946
|
pbx3a
|
pre-B-cell leukemia homeobox 3a |
| chr21_-_11996769 | 0.30 |
ENSDART00000143537
|
zgc:64106
|
zgc:64106 |
| chr11_-_669270 | 0.30 |
ENSDART00000172834
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr5_+_9377005 | 0.30 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr16_+_14029283 | 0.30 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr15_+_47386939 | 0.29 |
ENSDART00000128224
|
FO904873.1
|
|
| chr14_-_34633960 | 0.29 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr8_+_25299069 | 0.29 |
ENSDART00000114676
|
gstm.2
|
glutathione S-transferase mu tandem duplicate 2 |
| chr22_-_26558166 | 0.29 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr4_+_12931763 | 0.28 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr19_+_56351 | 0.28 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr8_-_7386978 | 0.28 |
ENSDART00000058321
|
lhfpl4b
|
LHFPL tetraspan subfamily member 4b |
| chr11_+_3254524 | 0.28 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr21_+_1381276 | 0.27 |
ENSDART00000192907
|
TCF4
|
transcription factor 4 |
| chr3_+_25216790 | 0.27 |
ENSDART00000156544
|
c1qtnf6a
|
C1q and TNF related 6a |
| chr8_-_46897734 | 0.27 |
ENSDART00000138125
|
hes2.2
|
hes family bHLH transcription factor 2, tandem duplicate 2 |
| chr12_-_35393211 | 0.27 |
ENSDART00000137139
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr9_+_55075526 | 0.27 |
ENSDART00000139555
ENSDART00000133864 |
gpr143
|
G protein-coupled receptor 143 |
| chr6_+_29923593 | 0.27 |
ENSDART00000169687
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr14_-_47882706 | 0.27 |
ENSDART00000188772
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr16_+_50741154 | 0.27 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr25_-_10565006 | 0.27 |
ENSDART00000130608
ENSDART00000190212 |
galn
|
galanin/GMAP prepropeptide |
| chr22_+_35275206 | 0.26 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr16_-_35975254 | 0.26 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr6_+_49082796 | 0.26 |
ENSDART00000182446
|
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr5_-_21044693 | 0.26 |
ENSDART00000140298
|
si:dkey-13n15.2
|
si:dkey-13n15.2 |
| chr1_-_9104631 | 0.26 |
ENSDART00000146642
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr23_-_46217134 | 0.25 |
ENSDART00000189477
ENSDART00000168352 |
CABZ01102528.1
|
|
| chr7_-_36358303 | 0.25 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr19_-_9472893 | 0.25 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr10_+_7182423 | 0.25 |
ENSDART00000186788
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr2_-_15041846 | 0.25 |
ENSDART00000139050
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr4_-_4535189 | 0.25 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr5_-_45651548 | 0.25 |
ENSDART00000097645
|
NPFFR2
|
neuropeptide FF receptor 2a |
| chr24_-_37877743 | 0.24 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr4_+_8376362 | 0.24 |
ENSDART00000138653
ENSDART00000132647 |
erc1b
|
ELKS/RAB6-interacting/CAST family member 1b |
| chr11_-_18799827 | 0.24 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr1_+_56229674 | 0.23 |
ENSDART00000127714
|
c3a.2
|
complement component c3a, duplicate 2 |
| chr3_-_54544612 | 0.23 |
ENSDART00000018044
|
angptl6
|
angiopoietin-like 6 |
| chr21_+_22985078 | 0.23 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr6_-_33707278 | 0.23 |
ENSDART00000188103
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr7_-_23848012 | 0.23 |
ENSDART00000146104
ENSDART00000175108 |
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr2_+_25378457 | 0.22 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr24_-_41267184 | 0.22 |
ENSDART00000063504
|
xylb
|
xylulokinase homolog (H. influenzae) |
| chr14_+_7140997 | 0.22 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr7_-_36358735 | 0.22 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr4_+_10616626 | 0.22 |
ENSDART00000067251
ENSDART00000143690 |
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_+_39401388 | 0.22 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr17_-_44584811 | 0.22 |
ENSDART00000165059
ENSDART00000165252 |
slc35f4
|
solute carrier family 35, member F4 |
| chr14_+_23076207 | 0.21 |
ENSDART00000161628
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr13_+_30912385 | 0.21 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr15_+_25681044 | 0.21 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr21_+_19635486 | 0.20 |
ENSDART00000185736
|
fgf10a
|
fibroblast growth factor 10a |
| chr12_+_316238 | 0.20 |
ENSDART00000187492
|
rcvrnb
|
recoverin b |
| chr21_+_26991198 | 0.20 |
ENSDART00000065397
|
fkbp2
|
FK506 binding protein 2 |
| chr22_+_1028724 | 0.20 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr14_+_36521553 | 0.20 |
ENSDART00000136233
|
TENM3
|
si:dkey-237h12.3 |
| chr22_+_12366516 | 0.19 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr23_-_28294763 | 0.19 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr1_+_50976975 | 0.19 |
ENSDART00000022290
ENSDART00000140982 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr11_+_27364338 | 0.19 |
ENSDART00000186759
|
fbln2
|
fibulin 2 |
| chr13_-_25719628 | 0.18 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr17_-_3303805 | 0.18 |
ENSDART00000169136
|
CABZ01007222.1
|
|
| chr14_+_30285613 | 0.18 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr7_-_23777445 | 0.18 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr2_+_28889936 | 0.18 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr14_+_39255437 | 0.18 |
ENSDART00000147443
|
diaph2
|
diaphanous-related formin 2 |
| chr15_-_21692630 | 0.18 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr24_-_37877978 | 0.17 |
ENSDART00000128574
|
tmem204
|
transmembrane protein 204 |
| chr3_+_60007703 | 0.17 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr8_-_54304381 | 0.17 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr9_-_21238159 | 0.17 |
ENSDART00000146764
ENSDART00000102143 |
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr2_-_51095743 | 0.17 |
ENSDART00000184646
|
THEM6
|
si:ch73-52e5.2 |
| chr19_-_24443394 | 0.17 |
ENSDART00000090825
|
thbs3b
|
thrombospondin 3b |
| chr19_-_46957968 | 0.17 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr19_+_37620342 | 0.17 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr15_-_32383529 | 0.17 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr5_-_8171625 | 0.17 |
ENSDART00000167643
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr14_+_14225048 | 0.17 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr15_+_29728377 | 0.16 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr14_+_8328645 | 0.16 |
ENSDART00000127494
|
nrg2b
|
neuregulin 2b |
| chr17_+_24446353 | 0.16 |
ENSDART00000140467
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr6_-_32726848 | 0.16 |
ENSDART00000155294
|
zc3h3
|
zinc finger CCCH-type containing 3 |
| chr19_-_24443867 | 0.16 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr8_-_14126646 | 0.16 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr18_+_5490668 | 0.16 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr18_+_50278858 | 0.15 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr23_-_32892441 | 0.15 |
ENSDART00000147998
|
plxna2
|
plexin A2 |
| chr3_-_61592417 | 0.15 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr5_+_37487687 | 0.15 |
ENSDART00000184659
|
CU984579.1
|
|
| chr4_+_5798223 | 0.15 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr18_+_48446704 | 0.15 |
ENSDART00000134817
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr12_-_6551681 | 0.15 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr11_-_37411492 | 0.15 |
ENSDART00000166468
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr25_-_19661198 | 0.15 |
ENSDART00000149641
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr2_+_51028269 | 0.15 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr17_-_37052622 | 0.15 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr16_-_43065164 | 0.15 |
ENSDART00000149431
|
sema4aa
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Aa |
| chr24_-_28711176 | 0.14 |
ENSDART00000105753
|
olfm3a
|
olfactomedin 3a |
| chr12_+_40905427 | 0.14 |
ENSDART00000170526
ENSDART00000185771 ENSDART00000193945 |
CDH18
|
cadherin 18 |
| chr2_+_5948534 | 0.14 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr20_+_1960092 | 0.14 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr8_+_25342896 | 0.14 |
ENSDART00000129032
|
CR847543.1
|
|
| chr23_+_45611649 | 0.14 |
ENSDART00000169521
|
dclk2b
|
doublecortin-like kinase 2b |
| chr12_+_4573696 | 0.14 |
ENSDART00000152534
|
si:dkey-94f20.4
|
si:dkey-94f20.4 |
| chr14_+_14224730 | 0.14 |
ENSDART00000180112
ENSDART00000184891 ENSDART00000174760 |
nlgn3a
|
neuroligin 3a |
| chr23_-_42093537 | 0.14 |
ENSDART00000122074
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr23_+_16620801 | 0.14 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr16_+_50972803 | 0.13 |
ENSDART00000178189
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr2_-_6548838 | 0.13 |
ENSDART00000052419
|
rgs18
|
regulator of G protein signaling 18 |
| chr22_-_29586608 | 0.13 |
ENSDART00000059869
|
adra2a
|
adrenoceptor alpha 2A |
| chr13_-_226393 | 0.13 |
ENSDART00000172677
|
rtn4b
|
reticulon 4b |
| chr8_+_7756893 | 0.13 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr2_-_53896300 | 0.13 |
ENSDART00000161221
|
capsla
|
calcyphosine-like a |
| chr13_+_27314795 | 0.13 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr14_-_30876708 | 0.13 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr6_+_19933763 | 0.13 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr22_+_15438872 | 0.13 |
ENSDART00000139800
|
gpc5b
|
glypican 5b |
| chr1_+_59538755 | 0.13 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr3_+_14339728 | 0.13 |
ENSDART00000184127
|
plppr2a
|
phospholipid phosphatase related 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-149i17.7_zic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.2 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 0.5 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.4 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.3 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.2 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.2 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 1.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.3 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.6 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.2 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.5 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.3 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.4 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.0 | 0.4 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.5 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.1 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.1 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.1 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.3 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.0 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.4 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 1.3 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.4 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.5 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.3 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.2 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |